ssDNA VIRUS EVOLUTION, PATHOGENESIS, and ANTI-CANCER POTENTIAL
Research summary:
We investigate the molecular biology of ssDNA viruses, with special emphasis in virus members of the Parvoviridae, to understand their evolution patterns, mechanisms underlying pathogenicity, and oncolytic potential against human cancer.
Pathogenicity.
In these studies, we combine mice infections with sequence analysis of the parvovirus Minute Virus of Mice (MVM) genetic variants arising at precise stages of the caused diseases. The evolutionary capacity of this virus in response to immune and adaptive pressures is monitored by genome sequencing. This allows at certain cases to localize the selected amino acid changes at defined functional domains of the 3-D capsid structure (Figure 1).
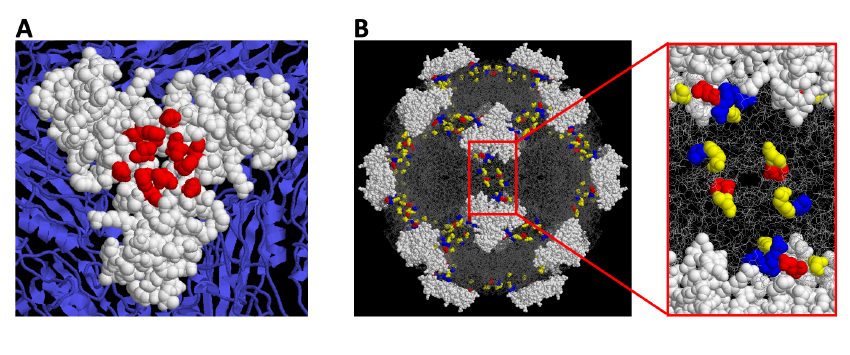
Figure 1. A Localization of Monoclonal antibody-resistant mutations in the parvovirus MVMi capsid. B. A constellation of residues at the 2fold axis of the MVM capsid contributes to the hemopoietic disease caused by the virus in scid mice.
Parvovirus MVM life cycle.
The preferent infection capacity of cancer cells (oncolysis) by MVM is being addressed by an in-depth analysis of the virus life cycle steps in human cancer cells, aimed at identifying regulators and precise molecular interactions at each major virus life cycle stage. Hence, we demonstrated that capsid assembly proceeds as trimeric intermediates that translocate through the nuclear envelope at the S phase of the cell cycle, and this nuclear transport competence relies on the phosphorylation at specific sites (Figure 2A) of the protein subunits mediated by the Raf-1 kinase (MAPK route) commonly deregulated in cancer cells (Figure 2B).
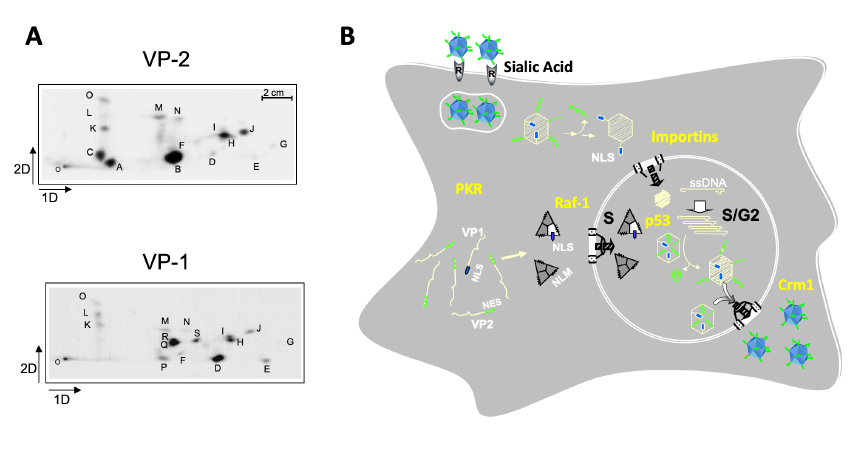
Figure 2. Major steps in the parvovirus MVM life cycle. A. Specific phosphorylation sites in the VP1 and VP2 capsid subunits. B. MVM life cycle regulators relevant for human cancer.
One aspect of MVM life-cycle regulation of particular interest for its oncolytic capacity is the dependence of S-phase on the transport of capsid subunits, and thus on the nuclear assembly of the virus (Figure 3). This coupling is relevant for understanding parvovirus pathogenesis and tropism for proliferative cells. Its disturbance may determine viral persistence in host tissues.
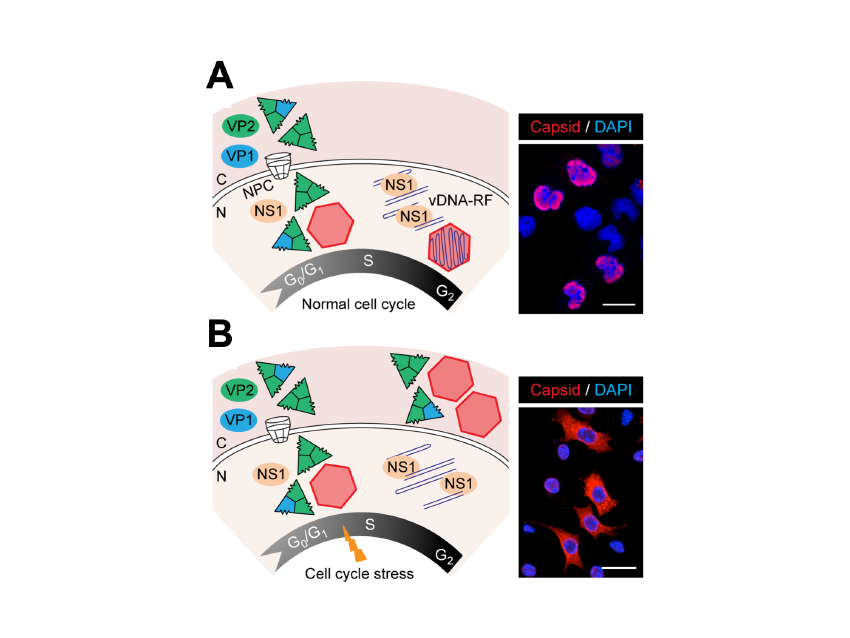
Figure 3. Time and functional coupling between cell cycle and parvovirus nuclear assembly.
Anti-cancer applications of the parvovirus MVM.
The anti-cancer therapeutic potential of the MVM was shown by its capacity to infect and eliminate cancer stem cells obtained from glioblastoma patients (the most aggressive brain tumor) implanted in the brain of rodent models (Figure 4). In neurospheres the MVM targets its cytotoxic action exclusively against those cells that have altered both innate responses and the central regulator p53 by mutation or aberrant phosphorylation, in a patient-dependent manner. These results raise hopes for a new personalized and biosafe medicine, based on viruses that selectively infect malignant stem cells, that could be used in therapies against this devastating type of cancer and against others carrying deregulated p53 signaling.
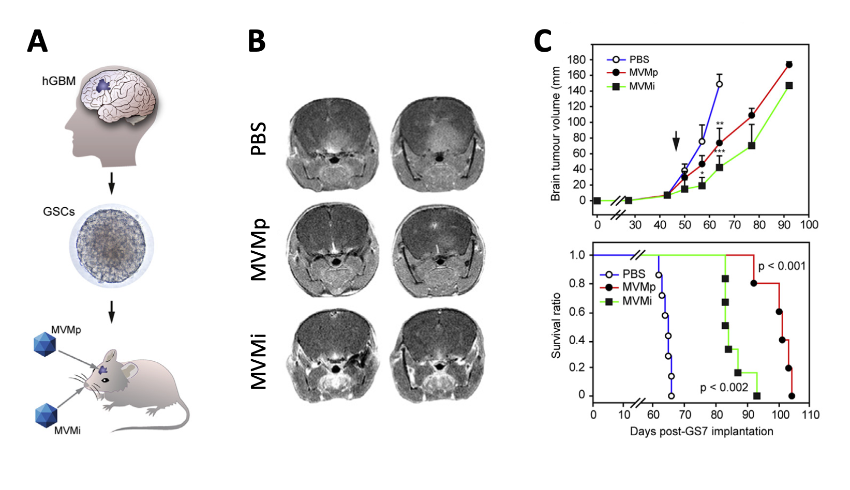
Figure 4. Therapeutic capacity of MVM against GSC-derived brain tumours in mice. A. Scheme of the methodology. B. MRI images of brain tumours at 60 d post-implantation. C. Tumour growth and Kaplan-Meyer survival analysis of the implanted mice.
Other ongoing approaches in our lab to enhance MVM oncolysis tackle re-targeting the virus to the neovascularization process required for tumor growth. We are engineering different domains of MVM capsid with heterologous peptides blocking VEGF (Figure 5A), to either induce antibodies that may reduce tumor growth (Figure 5B), or to drive the tropism of the virus specifically to VEGF-R expressing vascular cells supporting the tumor vascularization (Figure 5C).
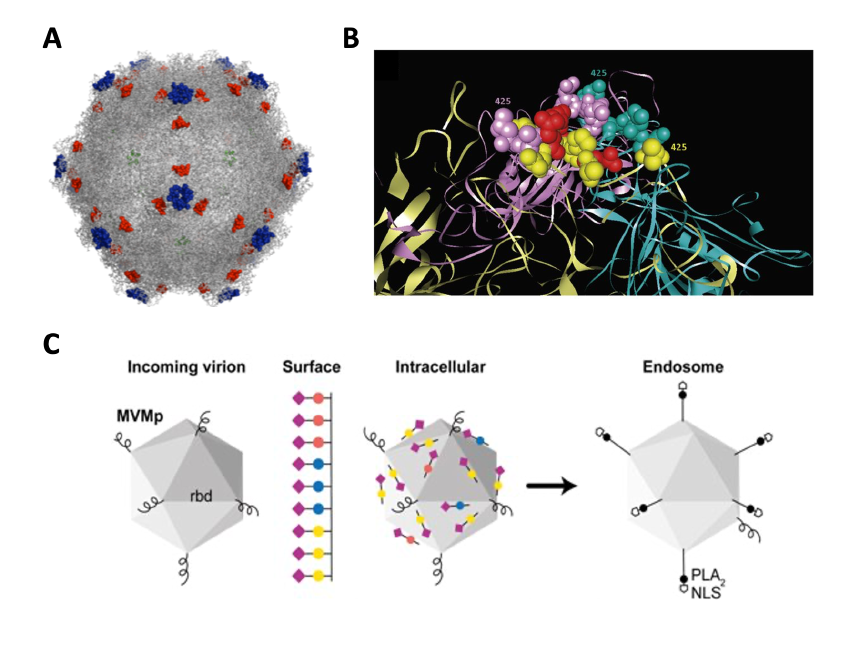
Figure 5. Manipulations of the MVM capsid with heterologous peptides. A. MVM capsid domains manipulated with VEGF-blocking peptides. B. The four loops that make up the spicule on the 3x symmetry axis replaced by peptides. C. The peptides help to understand the mechanism of sialic receptor-dependent virus entry.
MVM evolution confers advantageous anti-glioblastoma properties.
We are attempting to increase MVM tropism for cancer cells through multiple experimental strategies: (i). Under natural evolution in mice, MVM variants arise that contain amino acid changes in a “dimple” at the 2-fold axis of symmetry of the capsid. Some of them modulate the binding affinity for the sialic acid (sia) cell receptor and show improved infectivity of glioblastoma cells. The molecular basis of this enhanced onco-tropism is related to an important effect of the sia-type contacts on the endocytic traffic of the virion. (ii) We have developed serveral directed evolution strategies in vitro, based on the generation of MVM libraries by mutagenesis ( error-prone PCRs or saturation of precise codons) of the cellular receptor binding site and subsequent adaptation to glioblastoma cells through serial passages. The goal is to retarget MVM capsids toward sialoglycans preferentially expressed in glioblastoma cell lines. Some of the recombinant viruses engineered with the selected mutatnions (Fig. 6) replicate and propagate better than the wild type virus in the U373 glioblastoma cell line. (iii) Finally, we are fostering genetic recombination between the MVMp and MVMi strains by high multiplicity of co-infection of permissive cells. Oncotropic chimeric viruses emerged spontaneously in these cultures and are subjected to selection in U373 cells. One of the resulting viruses displays chimeric nonstrutural proteins and produce more viral genomes in U373 than the two parental strains, suggesting an enhanced oncotropism.
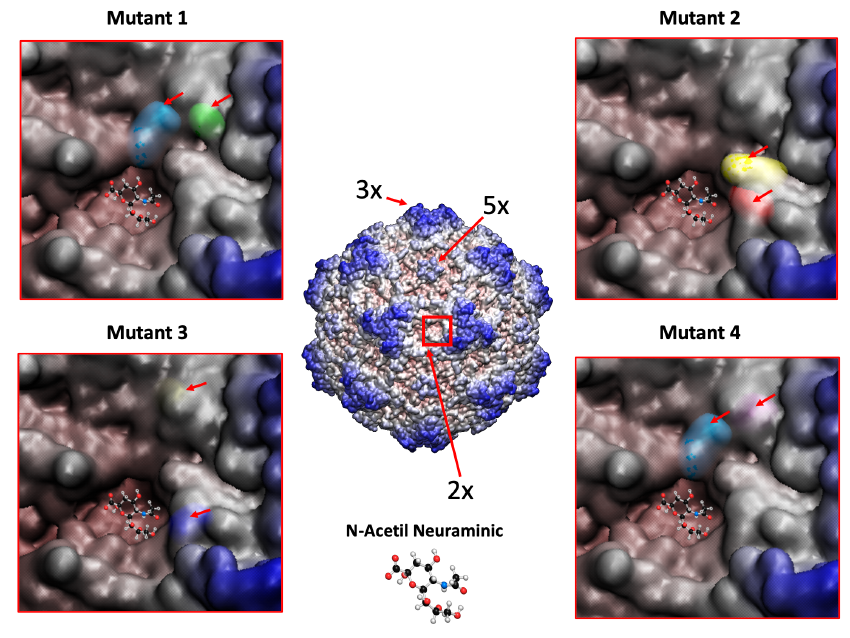
Figure 6. MVM mutants obtained by directed evolution towards a human glioblastoma cell line show amino acid changes nearby the dimple on the capsid surface. 3D structure of the MVM capsid with residues colored from red to blue depending on the distance to the center of the particle. The 3x (“spike”), 5x (“pore”) and 2x (“dimple”) axes of symmetry are indicated, as well as closed-up images of the latter region for four of these mutants with the changing residues indicated with an arrow.
Funding sources along recent years:
- “Re-targeting parvovirus infection to cellular processes determining human cancer”. PID2022-141799OB-I00. Ministerio de Innovación y Ciencia. Principal investigator: J.M. Almendral. Sep 2023-Ago 2026.

- “Directed parvovirus evolution aimed at human cancer therapy”. PID2019-111146RB-I00. Ministerio de Ciencia, Innovación y Universidades. Principal investigators: A. López-Bueno and J.M. Almendral. Jun 2020-Nov 2023.

- “Plataformas y modelos preclínicos para el abordaje multidisciplinar en COVID-19 y en respuesta a futuras pandemias”. COVTRAVI-19-CM. Coordinator: Manuel Fresno. Principal investigators: López-Bueno and J.M. Almendral. Feb 2022-Dec 2022.
- “Use of nonionizing radiation (modulation) to enhance cancer treatment with oncolytic viruses: effects on cell viability and signaling pathways in tumor models”. Paso Alto Biophysics and Biomedical Engineering S.L., Principal investigators: Y. Revilla and J.M. Almendral. Nov 2022-Dec 2023.

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Almendral del Río | José Mª | 224 | 4559 | jmalmendral(at)cbm.csic.es | Catedrático Universidad, GA |
| Fernández Llorente | Alejandro | 224 | 4559 | alejandro.fernandez(at)cbm.csic.es | Titulado Sup.de Actividades Técn. y Profes. GP1 |
| Flechoso Fernández | Gonzalo | 224 | 4559 | Estudiante TFM | |
| Gutiérrez Fombona | María | 224 | 4589 | maria.gutierrez(at)cbm.csic.es | Titulado Sup.de Actividades Técn. y Profes. GP1 |
| López Bueno | Alberto | 224 | 4589 | alopezbueno(at)cbm.csic.es | Profesor Titular Universidad, GA |
| Martínez Ortega | Jorge | 224 | 4589 | j.martinez(at)cbm.csic.es | Titulado Sup.de Actividades Técn. y Profes. GP1 |
| Ruiz Medina | Ana | 224 | 4559 | Estudiante TFG |
Publicaciones relevantes:
- Castellanos, M., Pérez, R., Rodríguez-Huete, A., Grueso, E., Almendral, J.M*., and Mateu, M.G*. 2013. A slender tract of glycines is required for translocation of protein VP2 N-terminal domain through the parvovirus MVM capsid channel to initiate infection. Biochem J,. 455, 87-94 (*, co-directors). https://doi.org/10.1042/BJ20130503
- Almendral, J.M. 2013. Assembly of simple icosahedral viruses. Subcell Biochem book series 68: 307-328. doi: https://doi.org/10.1007/978-94-007-6552-8_10
- Gil-Ranedo, , Hernando, E., Riolobos, L., Domínguez, C., Kann, M., and J.M. Almendral. 2015. The mammalian cell cycle regulates parvovirus nuclear capsid assembly. Plos Pathogens. 11;11(6):e1004920. https://doi.org/10.1371/journal.ppat.1004920.
- López-Bueno A., Rastrojo A., Peiró R., Arenas M., and A. Alcamí. 2015. Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Ecol. 24(19):4812-25. https://doi.org/10.1111/mec.13321. Cover of the Journal.
- López-Bueno A., Mavian C., Labella A.M., Castro D., Borrego J.J., Alcami A., and A. Alejo. 2016. Concurrence of Iridovirus, Polyomavirus, and a Unique Member of a New Group of Fish Papillomaviruses in Lymphocystis Disease-Affected Gilthead Sea Bream. Virol. 12;90(19):8768-79. https://doi.org/10.1128/JVI.01369-16
- Ros, , Bayat N., Wolfisberg R., and J.M. Almendral. 2017. Protoparvovirus Cell Entry. Viruses. 9, 313. https://doi.org/10.3390/v9110313.
- Gil-Ranedo J., Hernando E., Valle N., Riolobos L., Maroto B., and J.M. Almendral. 2018. Differential phosphorylation and n-terminal configuration of capsid subunits in parvovirus assembly. Virology. 518, 184–194. https://doi.org/10.1016/j.virol.2018.02.018.
- Parras-Moltó M., Rodríguez-Galet A., Suárez-Rodríguez P., and A. López-Bueno. 2018. Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses. Microbiome. 28;6(1):11. https://doi.org/10.1186/s40168-018-0507-3.
- Grueso, C., Sánchez-Martínez T., Calvo-López F.J., de Miguel N., Blanco-Menéndez M., Fernández-Estévez M., Elizalde J., Sánchez O., Kourani D., Martin A., Tato M., Guerra S., Andrés G., and J.M. Almendral. 2019. Antiangiogenic VEGF-Blocking Peptides Displayed on the Capsid of an Infectious Oncolytic Parvovirus: Assembly and Immune Interactions. J. Virol. 93, Issue 19 e00798-19. https://doi.org/10.1128/JVI.00798-19.
- Mavian C., López-Bueno A, Martín R, Nitsche A, Alcamí A. 2021. Comparative Pathogenesis, Genomics and Phylogeography of Mousepox. Viruses. 15;13(6):1146. https://doi.org/10.3390/v13061146 .
- Gil-Ranedo J, Gallego-García C, Almendral JM. 2021. Viral targeting of glioblastoma stem cells withpatient-specific genetic and post-translational p53 deregulations. Cell Reports. Sep 7;36(10):109673. https://doi.org/10.1016/j.celrep.2021.109673 .
- Calvo-López T, Grueso E, Sánchez-Martínez C, Almendral JM. Intracellular virion traffic to the endosome driven by cell type specific sialic acid receptors determines parvovirus tropism. Front Microbiol. Jan 23;13:1063706. https://doi.org/10.3389/fmicb.2022.1063706.
- Mietzsch M, Qiu J, Almendral JM and Söderlund-Venermo M (2023) Editorial: Parvoviruses: from basic research to biomedical and biotechnological applications. Microbiol. 14:1194926. https://doi.org/10.3389/fmicb.2023.1194926.
- López-Bueno A, Gil-Ranedo J, Almendral JM. 2024. Chapter 10: Assembly of small icosahedral viruses. Structure and Physics of Viruses. In press in Subcell Biochem book series.
Doctoral theses:
- Tania Calvo-López. 2021. “Ingeniería de dominios funcionales de la cápsida del parvovirus MVM con péptidos que bloquean VEGF: efectos en ensamblaje, inmunogenicidad y tropismo”. UAM. Director: JM Almendral.
- Carlos Gallego García. 2020. “Terapia de glioblastoma por el parvovirus MVM: implicación de p53 y su modulación por quimioterapia genotóxica”. UAM. Director: JM Almendral.
- Marcos Parras Moltó. 2019. “Estudio metagenómico de la comunidad de virus y de su interacción con la microbiota en la cavidad bucal humana”. UAM. Director: Alberto López Bueno.
- Nooshin Bayat. 2018. “Modificaciones de la cápsida de parvovirus en la entrada de células de cáncer e invasión evolutiva de genomas de primates/Parvovirus capsid modifications in cancer cell entry and evolutionary invasion of primate genomes”. UAM. Director: JM Almendral.




















