Replication of Bacteriophage ø29 DNA
Research summary:
For more than 50 years, our group has studied the molecular mechanisms of bacteriophage replication Φ29, from a multidisciplinary perspective. Under the scientific and personal leadership of Professor Margarita Salas, multiple biochemical and genetic aspects of the virus life cycle have been unraveled. These results supported the development of a number of biotechnological applications.
The loss of Dr. Salas is undoubtedly irreplaceable. However, we are determined to make the necessary effort, not only to maintain its legacy but also to complete the current research projects. The excellence and scientific rigor that Margarita instilled in us will help us to complete this task, which we consider is the best way in which his disciples can pay homage.
Obituario de Margarita Salas in Nature
Main current research lines.
- New aspects of DNA replication of bacteriophage Φ29 in vitro and in vivo.
This line of research includes a detailed study of the subcellular localization of viral replication in Bacillus subtilis, its natural host, as well as new advances in the molecular characterization of the virus's replicative machinery, through in vitro experiments. - Betatectivirus as a new model of replication of linear genomes initiated by terminal protein.
Tectiviruses are icosahedral bacterial viruses, without glue and containing an inner membrane under the capsid and can infect Gram-negative (Alfatectivirus) or Gram-positive (Beta- and Gammatectivirus) bacteria. This group of viruses has gained relevance in recent years, thanks to various studies on its ecological and evolutionary relevance. We have recently characterized viral DNA polymerase and the mechanism of protein-primed genome replication initiation of Bam35 virus, the model virus of the genus Betatectivirus. Currently, our work focuses on the characterization of viral DNA binding proteins, such as an a putative SSB (single stranded DNA binding protein), which may play an essential role in viral genome replication.
Additionally, in collaboration with Annika Gillis and Jacques Mahillon (Université Catholique de Louvain, Belgium), we are characterizing the replication mechanism of a new Betatectivirus. - New biotechnological activities and applications of viral DNA polymerases.
In the last 10 years, in collaboration with Miguel de Vega (CBMSO, CSIC), our laboratory has developed variants of Φ29 DNA polymerase with improved amplification efficiency and capacity (1) as well as with enhanced thermo-resistance (11). As a continuation of this price work, we are currently working on new variants of this DNA polymerase that allow the amplification of DNAs in different conditions.
The characterization of Bam35 DNA polymerase (B35DNAP), allowed us to find a new enzyme with possible biotechnological applications. Like the Φ29 DNA polymerase, B35DNAP replicates the DNA faithfully, processively and coupled to the displacement of the non-replicated strand. In addition, this new DNA polymerase is capable of replicating damaged DNA templates (TLS capability). We are currently working on obtaining engineered variants that allow amplifying reluctant DNA samples. - Global study of the virus-host protein interactome through the use of double hybrid coupled to analysis by mass sequencing.
The development of high-throughput methods allows the study of biological systems from a global point of view, in which each and every one of its elements are analyzed. For example, protein-protein interactions (PPI) in a given biological model can be comprehensively analyzed by modifying methods already known as the double yeast hybrid (Y2H). Through the application of this methodology, we are analyzing the PPIs for the first time in a virus-host model formed by a bacteriophage-bacterium, specifically, a system formed by the tempered Bam35 and its natural host Bacillus thuringiensis HER1410. This methodology will allow us to characterize in more detail the lineage and the virus-host relationships. - Encapsulation of transcription machinery.
In collaboration with Paco Monroy (Universidad Complutense de Madrid) and within the consortium: Nanobiocargo, we are currently working on the encapsulation of the molecular machinery necessary for the transcription of Φ29 DNA within lipid vesicles. We will study the energy needed for this process, as well as the heat that dissipates. For this purpose we will use microscopy techniques and optical tweezers.
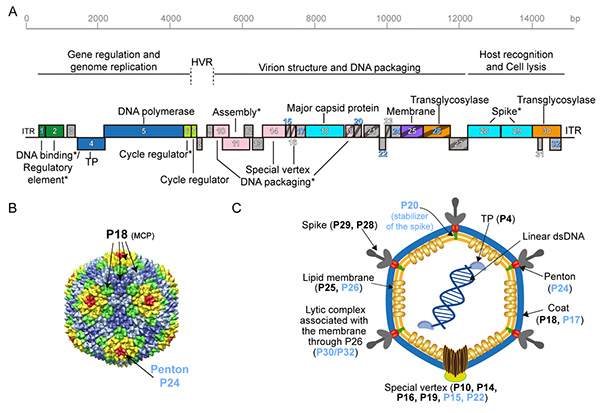
Figure 1. Bam35 virus genome and structure of the viral particle (Modified from Berjón-Otero et al., 2017). A. Genetic map of phage Bam35. The boxes correspond to the ORF predicted in the Bam35 genome. B. Bam35 virus capsid model, based on the crystallographic structure of the PRD1 Alphatectivirus. C. Proposed model for the Bam35 viral particle. Schematic representation of the Bam35 viral particle in which the proposed or known location of each of its proteins is indicated.

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Lechuga Mateo | Ana | 409 | 4463 | a.lechuga(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Muruzabal Galarza | Ane | 409 | 4463 | Estudiante TFM | |
| Ordoñez Cencerrado | Carlos David | 409 | 4463 | cordonez(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Prado Díaz | Alicia del | 409 | 4463 | adelprado(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Redrejo Rodríguez | Modesto | 409 | 4463 | mredrejo(at)cbm.csic.es | Estancia/Investigador |
Relevant publications:
- de Vega, M., et al., Improvement of Φ29 DNA Polymerase Amplification Performance by Fusion of DNA Binding Motifs. PNAS, 2010. 107(38): p. 16506-11. DOI: 10.1073/pnas.1011428107
- Berjón-Otero, M., et al., DNA Polymerase from Temperate Phage Bam35 Is Endowed with Processive Polymerization and Abasic Sites Translesion Synthesis Capacity. PNAS, 2015. 112(27): p. E3476-84. DOI: 10.1073/pnas.1510280112
- Mojardin, L. and M. Salas, Global Transcriptional Analysis of Virus-Host Interactions between Phage Phi29 and Bacillus Subtilis. J Virol, 2016. 90(20): p. 9293-304. DOI: 10.1128/JVI.01245-16
- Berjón-Otero, M., et al., Disclosing Early Steps of Protein-Primed Genome Replication of the Gram-Positive Tectivirus Bam35. NAR, 2016. 44(20): p. 9733-9744. DOI: 10.1093/nar/gkw673
- Berjón-Otero, M., et al., Bam35 Tectivirus Intraviral Interaction Map Unveils New Function and Localization of Phage Orfan Proteins. J Virol, 2017. 91(19). DOI: 10.1128/JVI.00870-17
- Redrejo-Rodríguez, M., et al., Primer-Independent DNA Synthesis by a Family B DNA Polymerase from Self-Replicating Mobile Genetic Elements. Cell Rep, 2017. 21(6): p. 1574-1587. DOI: 10.1016/j.celrep.2017.10.039
- van Nies, P., et al., Self-Replication of DNA by Its Encoded Proteins in Liposome-Based Synthetic Cells. Nat Commun, 2018. 9(1): p. 1583. DOI: 10.1038/s41467-018-03926-1
- del Prado, A., et al., Noncatalytic Aspartate at the Exonuclease Domain of Proofreading DNA Polymerases Regulates Both Degradative and Synthetic Activities. PNAS, 2018. 115(13): p. E2921-E2929. DOI: 10.1073/pnas.1718787115
- del Prado, Rodríguez I, Lázaro JM, Moreno-Morcillo M de Vega M, Salas M (2019). New insights into the coordination between the polymerisation and 3´-5´exonuclease activities in f29 DNA polymerase. Scientific Reports. 2019 Jan 29;9(1):923. DOI: 10.1038/s41598-018-37513-7
- de la Torre, I., et al., Tyrosines Involved in the Activity of Phi29 Single-Stranded DNA Binding Protein. PLoS One, 2019. 14(5): p. e0217248. DOI: 10.1371/journal.pone.0217248
- del Prado, A., et al., The Loop of the TPR1 Subdomain of Phi29 DNA Polymerase Plays a Pivotal Role in Primer-Terminus Stabilization at the Polymerization Active Site. Biomolecules, 2019. 9(11). DOI: 10.3390/biom9110648
Doctoral theses:
- Mónica Berjón Otero (UAM, 2017) Caracterización molecular del bacteriófago BAM35. Mecanismo de replicación del DNA y estudio del interactoma proteico. Directors: Margarita Salas y Modesto Redrejo Rodríguez.
- Eugenia Santos Del Río (UAM, 2017) Papel del motivo LExE de las DNA polimerasas que inician con proteína terminal en la interacción con el nucleótido entrante. Estabilización de los sustratos en el centro activo de polimerización mediada por el subdominio TPR1. Directors: Margarita Salas y Miguel de Vega.
Patent applications:
- Variantes de la ADN Polimerasa del Bacteriófago Phi29 con termoactividad mejorada. Solicitud de Patente ES201531910A (2015), CSIC y CRG.
- Primer-Independent DNA Polymerases and Their Use for DNA Synthesis, Solicitud de pantente ES201731236A (2017), CSIC e Instituto Pasteur




















