TCR domains in T cell differentiation and (patho)physiological and therapeutic responses
Research summary:
T cells use their T cell receptor (TCR) to continuously scan Major Histocompatibility Complex (MHC) molecules presenting the immense repertoire of peptides derived from the proteome present in the host. Presentation of pathogen-derived peptides gives rise to beneficial immune responses, while responses against peptides derived from the host itself can lead to auto-immune manifestations. Our research focuses on understanding how the organization of the TCR into nanoclusters allows T cells to become activated upon recognition of only a few peptide-bound MHC molecules. We use transgenic mouse models to study the role of TCR nanoclusters during early T cell differentiation, where a selection process takes place that filters out potentially autoimmune T cells, and to understand how this organization impacts on the ability to mount protective memory and undesired autoimmune responses. We also develop approaches to improve T cell responses against cancer, focusing on the signaling domains of recombinant tumor-specific receptors. Our research shows that the identity and topology of TCR-derived signaling domains of these recombinant receptors determines their efficiency with respect to activating the T cells. These results have a direct implication for improving cellular cancer immunotherapy. They also provide new inroads into understanding the mechanisms underlying TCR function, which we are currently following-up upon. In a third, clinically-oriented project, we aim to generate T cells expressing Chimeric Antigen Receptors (CAR-T cells) recognizing Acute Myeloid Leukemia (AML) blasts. Cellular Immunotherapy against this type of cancer has been severely hampered by unacceptable toxicity against the patient’s myeloid precursors that express the same antigen as the ones targeted on the AMLs. As no better target antigens have been defined, we are generating CAR-T cells that use a logical gate formed by an activating CAR recognizing the shared antigen on precursors and AMLs and an inhibitory immune receptor that recognizes a ligand only expressed by the myeloid precursors. If successful, this concept can be applied to other type of cancers using appropriate combinations of activating and inhibiting receptors.
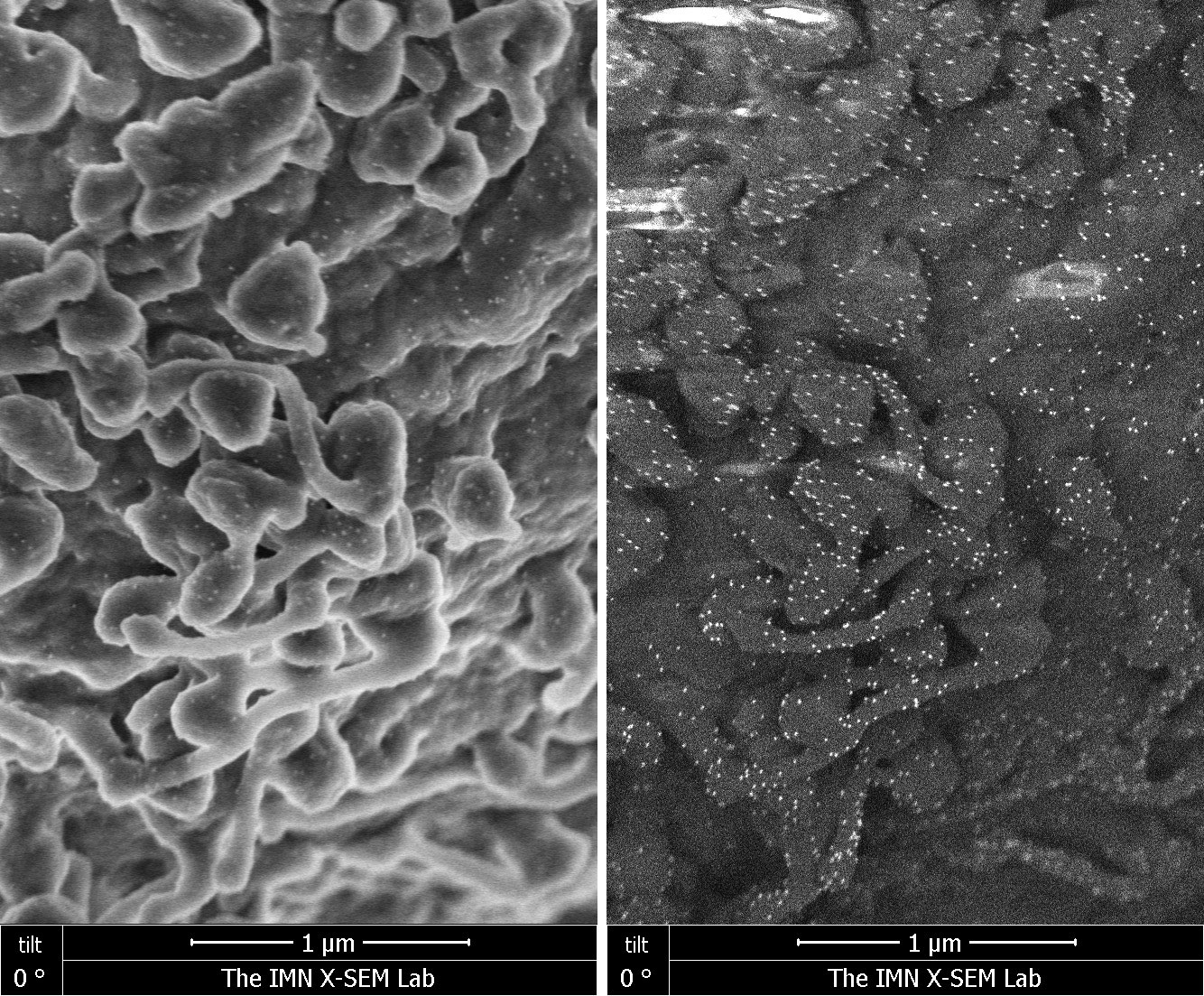
Scanning electron microscopy images (left panel: secondary electrons; right panel: back scatter electrons) of the Jurkat human T cell leukemia labeled with an antibody against the TCR complex and 10nm-gold conjugated protein A (white dots). Images obtained in collaboration with Drs. Gonzalez and San Pablo of the Institute of Micro and Nanotechnology (CSIC).

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Gutiérrez Pliego | María | 222 | 4682 | Estudiante | |
| Horndler Gil | Lydia Begoña | 221 | 4600 | lydia.horndler(at)cbm.csic.es | Investigador Indef. GP1 |
| Rodríguez Moraga | María | 222 | 4682 | Estudiante TFG | |
| Serrano Pérez | Irene | 221 | 4682 | iserrano(at)cbm.csic.es | M3 Predoc.formación |
| Van Santen | Hisse Martien | 221 | 4682 | hvansanten(at)cbm.csic.es | E.Científicos Titulares de Organismos Públicos de Investigación |
Relevant publications:
- Horndler, L, Delgado P, Abia D, Balabanov I, Martínez‐Fleta P, Cornish G, Llamas MA, Serrano‐Villar S, Sánchez‐Madrid F, Fresno M, van Santen HM, Alarcón B (2021). Flow cytometry multiplexed method for the detection of neutralizing human antibodies to the native SARS‐CoV‐2 spike protein. EMBO Mol Med 13: e13549; doi: 10.15252/emmm.202013549
- Martín-Leal A, Blanco R, Casas J, Sáez ME, Rodríguez-Bovolenta E, de Rojas I, Drechsler C, Real LM, Fabrias G, Ruíz A, Castro M, Schamel WWA, Alarcón B, van Santen HM, Mañes S (2020). CCR5 deficiency impairs CD4+ T cell memory responses and antigenic sensitivity through increased ceramide synthesis. EMBO J 39: e104749; doi: 10.15252/embj.2020104749
- Mirones I, Moreno L, Patiño-García A, Lizeaga G, Moraleda JM, Toribio ML, Pérez-Martínez A, en representación del Grupo de Inmunoterapia y Terapias Avanzadas de la Sociedad Española de Hematología y Oncología Pediátricas (24/26) (2020). Inmunoterapia con células CAR-T en hematooncología pediátrica. An Pediatr (Barc) 93: 59.e1-59.e10; doi: 10.1016/j.anpedi.2019.12.014.
- Martínez-Riaño A, Bovolenta ER, Boccasavia VL, Ponomarenko J, Abia D, Oeste CL, Fresno M, van Santen HM*, Alarcon B*. (2019). RRAS2 shapes the TCR repertoire by setting the threshold for negative selection. J. Exp. Med. 216, 2427-2447 (*: co-corresponding authors).
- Castro M*, van Santen HM*, Ferez M, Alarcón B, Lythe GD, Molina-Paris C (2014). Receptor Pre-Clustering and T cell Responses: Insights into Molecular Mechanisms. Front. Immunol. 5, 132; doi: 10.3389/fimmu.2014.00132 (*: co-corresponding authors).
- Férez M, Castro M, Alarcon B*, van Santen HM* (2014). Cognate peptide-MHC complexes are expressed as tightly apposed nanoclusters in virus-infected cells to allow TCR crosslinking. J. Immunol. 192, 52-58 (*: co-corresponding authors).
- Fiala G, Rejas MT, Schamel WW, van Santen HM (2013). Visualization of TCR Nanoclusters via Immunogold Labeling, Freeze-Etching, and Surface Replication. Methods in Cell Biology 117, 391 - 410.
- Bains I, van Santen HM, Seddon B, Yates AW (2013). Models of self-peptide sampling by developing T cells identify candidate mechanisms of thymic selection. PLoS Computational Biology 9 - 7, pp. e1003102.
- Kumar R, Ferez M, Swamy M, Arechaga I, Rejas MT, Valpuesta JM, Schamel WW, Alarcon B*, van Santen HM* (2011). Increased Sensitivity of Antigen-Experienced T Cells through the Enrichment of Oligomeric T Cell Receptor Complexes. Immunity 35, 375-387 (*: co-corresponding authors).
Book chapters and reviews:
- Alarcón B, van Santen HM (2016). T Cell Receptor Triggering. In: Ralph A Bradshaw and Philip D Stahl (Editors-in-Chief), Encyclopedia of Cell Biology, Vol 3, Functional Cell Biology, pp. 650-659. Waltham, MA: Academic Press; doi:10.1016/B978-0-12-394447-4.30097-9
- Martín-Blanco N, van Santen HM, Alarcón B (2016). TCR Signaling: Proximal Signaling. In: Ratcliffe, M.J.H. (Editor in Chief), Encyclopedia of Immunobiology, Vol. 3, pp. 1–8. Oxford: Academic Press http://dx.doi.org/10.1016/B978-0-12-374279-7.11002-1




















