Human immunodeficiency virus reverse transcriptase and antiretroviral therapy
Research summary:
Infections caused by human immunodeficiency viruses type 1 and type 2 (HIV-1 and HIV-2, respectively) constitute a major burden to human health worldwide. Despite significant advances in antiretroviral therapy HIV still causes 1.6 million deaths each year. The HIV genome is composed of two copies of single-stranded RNA. The viral reverse transcriptase (RT) is responsible for the replication of the HIV genome. RT inhibitors constitute the backbone of the most popular and effective therapies.
For years, our efforts have been directed towards achieving two major goals: (1) the elucidation of molecular mechanisms involved in RT inhibitor resistance; and (2) understanding the role of different amino acids in the nucleotide specificity of HIV-1 and HIV-2 RTs, as well as in their fidelity of DNA synthesis. We have engineered HIV-1 RT variants with increased thermal stability and high fidelity that are currently marketed as biotechnological tools for many applications, covering basic and translational research. Future challenges involve the development of RTs with high nucleic acid binding affinity suitable for RNA amplification from single cells.
HIV-2 shows natural resistance to nonnucleoside RT inhibitors (NNRTIs) and several protease inhibitors. Nucleoside RT inhibitors (NRTIs) constitute the backbone of therapies against HIV. However, HIV-1 and HIV-2 show different mutational pathways of NRTI resistance. We have recently demonstrated that HIV-2 RT residues Met-73 and Ile-75 restrict the development of M41L, D67N, K70R and T215Y (known as thymidine analogue resistance mutations). The reported findings explain why thymidine analogue resistance mutations are rarely observed in treated HIV-2-infected patients.
On the other hand, prevalence of drug-resistant HIV strains is increasing in less developed countries. Therefore, we anticipate an increasing interest in unexploited targets of antiretroviral intervention. In this context, RNase H activity and inhibition, as well as host factors that could eventually block HIV-1 replication are also important topics of research in our laboratory.
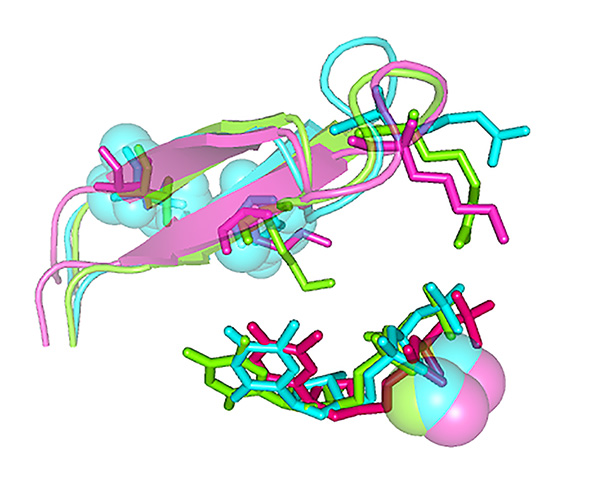
Comparison of the structural models of wild-type and mutant HIV-1 RTs D67N/K70R and D67N/K70R/K73M, complexed with double-stranded DNA and an incoming dNTP. View of the β3-β4 hairpin loop, the incoming dNTP and the two Mg2+ ions, highlighting the effects of Met-73 on the interaction of Arg-70 with the dNTP. For details see Álvarez et al. (2018) J. Biol. Chem., 293, 2247-2259.
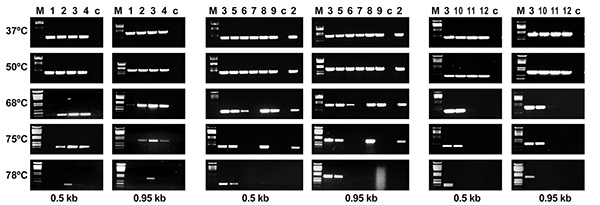
RT-PCR assays showing the effect of the temperature on the cDNA synthesis catalyzed by engineered recombinant HIV-1 group O RT variants (Matamoros et al. 2013; Biochemistry 52, 9318).

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Caz Calvo | Rubén del | 124 | 4523 | Estudiante TFM | |
| Estévez Barcia | Rebeca | 124 | 4523 | Estudiante TFG | |
| Martínez del Río | Javier | 124 | 4523 | javier.martinez(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Menéndez Arias | Luis | 124 | 4494 | lmenendez(at)cbm.csic.es | E. Profesores de Investigación de Organismos Públicos de Investigación |
| Rello García | Nuria | 124 | 4494/4523 | Estudiante TFM |
Relevant publications:
- Menéndez-Arias, L. (2013) Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antiviral Res. 98, 93-120; doi:10.1016/j.an-tiviral.2013.01.007.
- Álvarez, M., Barrioluengo, V., Afonso-Lehmann, R.N., Menéndez-Arias, L. (2013) Altered error specificity of RNase H-deficient HIV-1 reverse transcriptases during DNA-dependent DNA synthesis. Nucleic Acids Res. 41, 4601-4612; doi:10.1093/nar/gkt109.
- Matamoros, T., Barrioluengo, V., Abia, D., Menéndez-Arias, L. (2013) Major groove binding track residues of the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase enhance cDNA synthesis at high temperatures. Biochemistry 52, 9318-9328; doi:10.1021/bi401390x.
- Betancor, G., Álvarez, M., Marcelli, B., Andrés, C., Martínez, M.A., Menéndez-Arias, L. (2015) Effects of HIV-1 reverse transcriptase connection subdomain mutations on polypurine tract removal and initiation of (+)-strand DNA synthesis. Nucleic Acids Res. 43, 2259-2270.
- Álvarez, M., Sebastián-Martín, A., García-Marquina, G., Menéndez-Arias, L. (2017) Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases. Sci. Rep. 7, 44834. doi:10.1038/srep44834.
- Álvarez, M., Nevot, M., Mendieta, J., Martínez, M.A., Menéndez-Arias, L. (2018) Amino acid residues in HIV-2 reverse transcriptase that restrict the development of nucleoside analogue resistance through the excision pathway. J. Biol. Chem. 293, 2247-2259; doi:10.1074/jbc.RA117.000177.
- Sebastián-Martín, A., Barrioluengo, V., Menéndez-Arias, L. (2018) Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases. Sci. Rep. 8, 627; doi:10.1038/s41598-017-18974-8.
- Sun, L., Gao, P., Dong, G., Zhang, X., Cheng, X., Ding, X., Wang, X., Daelemans, D., De Clercq, E., Pannecouque, C., Menéndez-Arias, L., Zhan, P., Liu, X. (2018) 5-Hydroxypyrido[2,3-b]pyrazin-6(5H)-one derivatives as novel dual inhibitors of HIV-1 reverse transcriptase-associated ribonuclease H and integrase. Eur. J. Med. Chem. 155, 714-724; doi:10.1016/j.ejmech.2018.06.036.
- Luczkowiak, J., Matamoros, T., Menéndez-Arias, L. (2018) Template-primer binding affinity and RNase H cleavage specificity contribute to the strand transfer efficiency of HIV-1 reverse transcriptase. J. Biol. Chem. 293, 13351-13363; doi:10.1074/jbc.RA118.004324.
- Tramontano, E., Corona, A., Menéndez-Arias, L. (2019) Ribonuclease H, an unexploited target for antiviral intervention against HIV and hepatitis B virus. Antiviral Res. 171, 104613; doi: 10.1016/j.antiviral.2019.104613.




















