Bacterial cell envelope during preseptal growth
A major and essential structural component of the cell envelope of most bacteria is the peptidoglycan sacculus, and its synthesis is by far the most antibiotic-targeted process. In Gram-negative bacteria, the thin and net-like peptidoglycan layer is surrounded by an asymmetric and hydrophobic lipid bilayer (the outer membrane), which acts as a permeability barrier against external agents like many clinically used antibiotics. During the bacterial cell cycle both peptidoglycan and membrane biogenesis machineries are coordinated and regulated to ensure the robust growth of the cell envelope and the viability of the cell. Contrary to cell elongation and cell division, the transition between both stages called preseptal growth - synthesis of cell envelope at the division site before septation - remains poorly characterized.
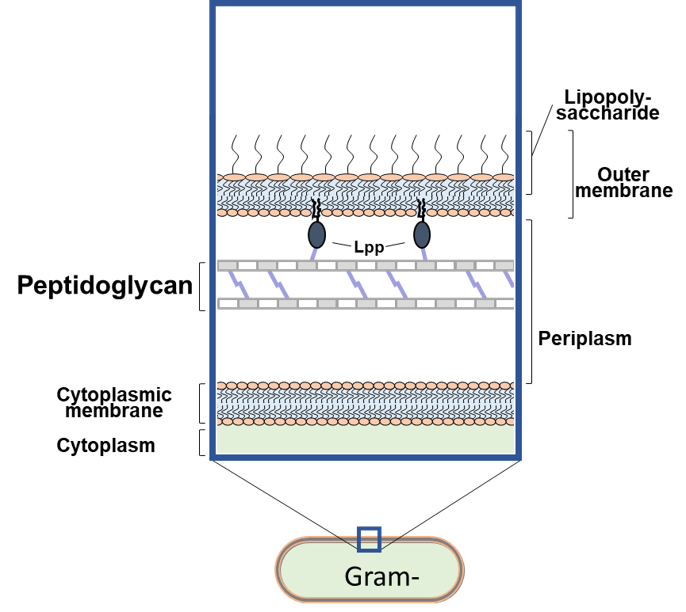
Figure 1. Schematic structure of the bacterial cell envelope in Gram-negative bacteria.
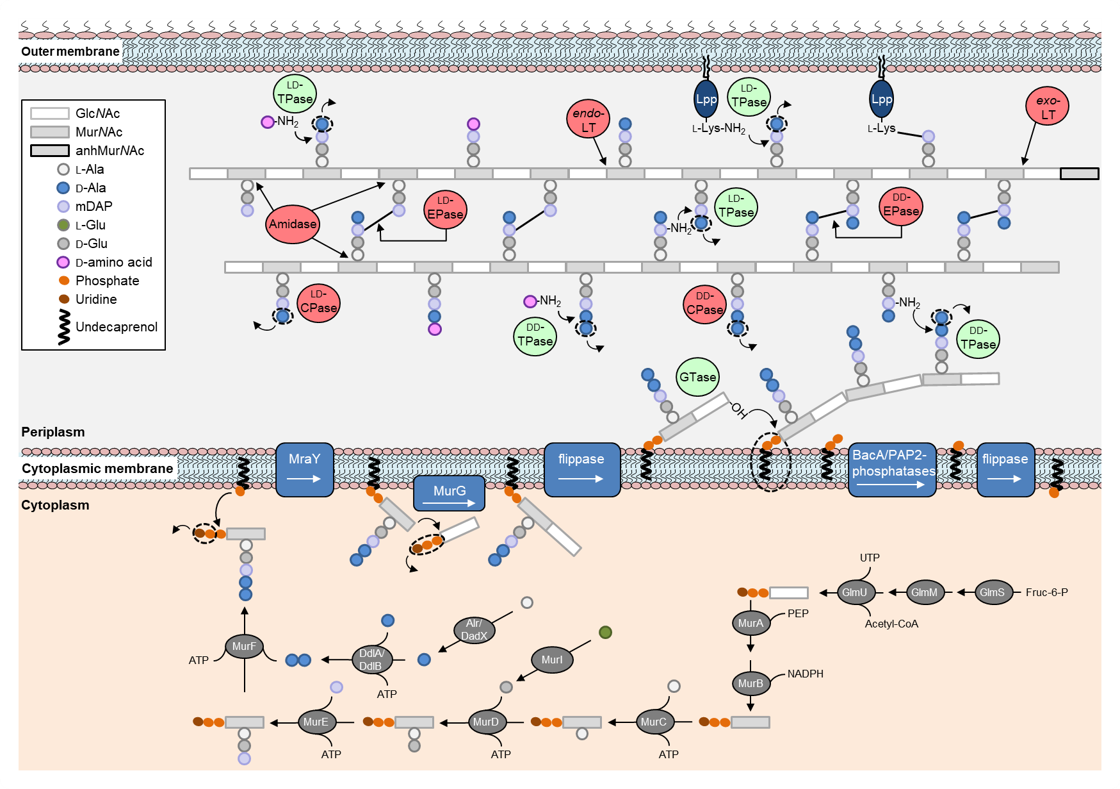
Figure 2. Synthetic and hydrolytic reactions occurring during the synthesis and incorporation of new peptidoglycan in Escherichia coli.
The research in the laboratory focuses on understanding the molecular mechanisms regulating the biogenesis of the bacterial cell envelope of Gram-negative bacteria, using gastrointestinal pathogenic organisms. Using a multidisciplinary approach combining genetics, biochemistry, cell biology and different microscopy techniques we aim to identify and characterize the protein interactions and their impact on the peptidoglycan enzymatic activities, the mechanical and structural properties of the cell envelope, and the impact on virulence.
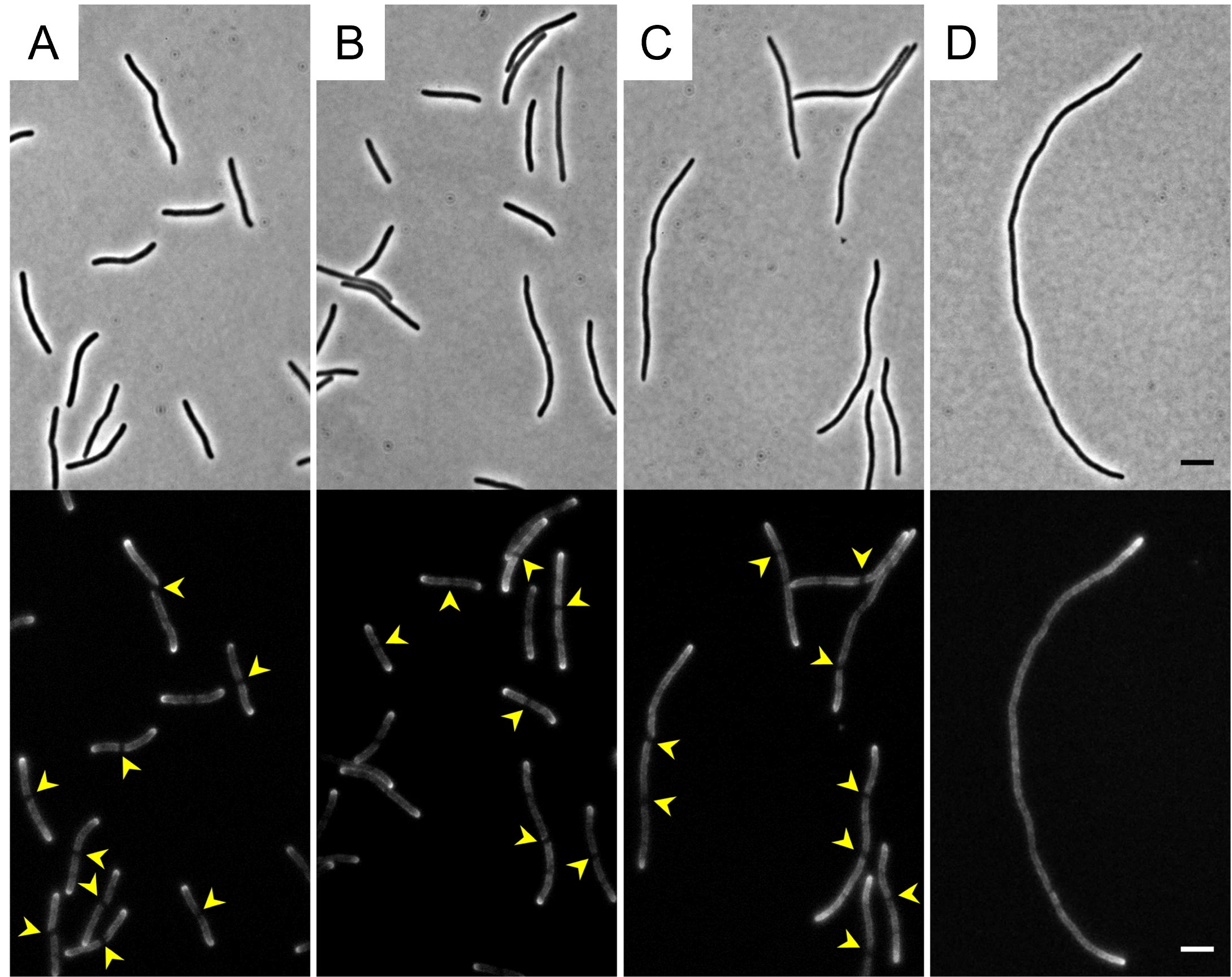
Figure 3. Preseptal peptidoglycan synthesis requires FtsN and ZipA. Phase contrast and HADA fluorescent images for detection of preseptal PG synthesis in the parental strain ftsAE124A (A), ZipA-depleted cells (B), FtsN-depleted cells (C) and ZipA- and FtsN-depleted cells (D). The non-fluorescently labelled preseptal PG synthesis bands are indicated by yellow arrows. Scale bars represent 5 µm.
| Apellidos | Nombre | Laboratorio | Ext.* | Categoría profesional | |
|---|---|---|---|---|---|
| Ballesteros San José | Daniel | 107 | 4530 | dballesteros(at)cbm.csic.es | M3 Predoc.formación |
| Belloso Casuso | Aitana | 107 | 4530 | abelloso(at)cbm.csic.es | Titulado Sup.de Actividades Técn. y Profes. GP1 |
| Bermejo López | Luis Fernando | 107 | 4530 | Estudiante TFG | |
| Lemus Aguilar | Luis Fernando | 107 | 4530 | lflemus(at)cbm.csic.es | M3 Predoc.formación |
| Pazos Don Pedro | Manuel | 107 | 4506 | mpazos(at)cbm.csic.es | Investigador Doctor |
20. Mamou G., Corona F., Cohen-Khait R., Housden N. G., Yeung V., Sun D., Sridhar P., Pazos M., Knowles T. J., Kleanthous C. and Vollmer W. (2022). Peptidoglycan maturation controls outer membrane protein assembly. Nature 606, 953-959. DOI: 10.1038/s41586-022-04834-7
19. Wacnik K., Rao V. A., Chen X., Lafage L., Pazos M., Booth S., Vollmer W., Hobbs J. K., Lewis R. J. and Foster S. J. (2022). Penicillin-Binding Protein 1 (PBP1) of Staphylococcus aureus has multiple essential functions in cell division. mBio 13:e00669-22. DOI: 10.1128/mbio.00669-22
18. Sassine J., Pazos M., Breukink E. and Vollmer W. (2021). Lytic transglycosylase MltG cleaves in nascent peptidoglycan and produces short glycan strands. Cell Surf 7:100053. DOI: 10.1016/j.tcsw.2021.100053
17. Pazos M. and Vollmer W. (2021). Regulation and function of class A Penicillin-binding proteins. Curr Opin Microbiol 60:80-87. DOI: 10.1016/j.mib.2021.01.008
16. Pazos M.*, Peters K., Boes A., Safaei Y., Kenward C., Caveney N.A., Laguri C., Breukink E., Strynadka N. C. J., Simorre, J.P., Terrak M and Vollmer W*. (2020). SPOR proteins are required for functionality of class A Penicillin-binding proteins in Escherichia coli. mBio 11:e02796-20. DOI: 10.1128/mBio.02796-20 *(co-corresponding author)
15. Banzhaf M., Yau HC., Verheul J., Lodge A., Kritikos G., Mateus A., Cordier B., Hov AK., Stein F., Wartel M., Pazos M., Solovyova AS., Breukink E., van Teeffelen S., Savitski MM., den Blaauwen T., Typas A. and Vollmer W. (2020). Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J e102246. DOI: 10.15252/embj.2019102246
14. Peters K., Pazos M., VanNieuwenhze MS. and Vollmer W. (2019). Optimized Protocol for the Incorporation of FDAA (HADA Labeling) for in situ Labeling of Peptidoglycan. Bio-protocol 9(15): e3316. DOI: 10.21769/BioProtoc.3316.
13. Pazos M.*, Peters K. (2019) Peptidoglycan. In: Kuhn A. (eds) Bacterial Cell Walls and Membranes. Subcellular Biochemistry, Springer, Cham. 92:127-168. DOI: 10.1007/978-3-030-18768-2_5 *(corresponding author)
12. Pazos M., Peters K., Casanova M., Palacios P., VanNieuwenhze MS., Breukink E., Vicente M. and Vollmer W. (2018). Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat Comm 9:5090. DOI: 10.1038/s41467-018-07559-2
11. Karinou E., Schuster CF., Pazos M., Vollmer W. and Gründling A. (2018). Inactivation of the monofunctional peptidoglycan glycosyltransferase SgtB allows Staphylococcus aureus to survive in the absence of lipoteichoic acid. J Bacteriol 201:e00574-18. DOI: 10.1128/JB.00574-18
10. Peters K., Pazos M., Edoo Z., Hugonnet JE., Martorana AM., Polissi A., VanNieuwenhze MS., Arthur M. and Vollmer W. (2018). Copper inhibits peptidoglycan LD-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc Natl Acad Sci U S A 115:10786-10791. DOI: 10.1073/pnas.1809285115.
9. Geiger T., Pazos M., Lara-Tejero M., Vollmer W. and Galán JE. (2018). Peptidoglycan editing by a specific LD-transpeptidase controls the muramidase-dependent secretion of typhoid toxin. Nat Microbiol 3:1243-1254. DOI: 10.1038/s41564-018-0248-x
8. Klein TA., Pazos M., Surette MG., Vollmer W. and Whitney JC. (2018). Molecular basis for immunity protein recognition of a type VII secretion system exported antibacterial toxin. J Mol Biol 430:4344-4358. DOI: 10.1016/j.jmb.2018.08.027
7. Pazos M., Otten C. and Vollmer W. (2018). Bacterial cell wall precursor phosphatase assays using thin-layer chromatography (TLC) and high pressure liquid chromatography (HPLC). Bio-protocol 8(6): e2761. DOI: 10.21769/BioProtoc.2761.
6. Whitney JC., Peterson SB., Kim J., Pazos M., Verster AJ., Radey MC., Kulasekara HD., Ching MQ., Bullen NP., Bryant D., Goo YA., Surette MG., Borenstein E., Vollmer W. and Mougous JD. (2017). A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. eLife 6:e26938 DOI:10.7554/eLife.26938
5. Pazos M., Peters K. and Vollmer W. (2017). Robust peptidoglycan growth by dynamic and variable multi-protein complexes. Curr Opin Microbiol 36:55-61 (cover picture of the issue)
4. Pazos M., Casanova M., Palacios P., Margolin W., Natale P. and Vicente M. (2014). FtsZ placement in nucleoid-free bacteria. PLoS One 9(3):e91984 DOI:10.1371/journal.pone.0091984
3. Natale P., Pazos M. and Vicente M. (2013). The Escherichia coli divisome: born to divide. Environ Microbiol 15(12):3169-3182 DOI:10.1111/1462-2920.12227
2. Pazos M., Natale P. Margolin W. and Vicente M. (2013). Interactions among the early Escherichia coli divisome proteins revealed by Bimolecular Fluorescence Complementation (BiFC). Environ Microbiol 15(12):3282-3291 DOI:10.1111/1462-2920.12225
1. Pazos M., Natale P. and Vicente M. (2013). A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J Biol Chem 288:3219-3226. DOI:10.1074/jbc.M112.434944




















