Programa Científico
Dinámica y función del genoma
GRUPO DE INVESTIGACIÓN
Iniciación interna de la traducción en mRNAs eucarióticos

Encarna Martinez-Salas
Our team aims to understand the interplay of RNA and RNA-binding proteins (RBPs) in translation control. We identified the dimerization and the non-canonical RNA-binding domain of Gemin5, a multifunctional RBP involved in splicing and translation, providing crucial information to investigate the processes disrupted by Gemin5 biallelic mutations reported in patients suffering neurological disorders
Investigación
Our aims are focused to understand the principles guiding alternative mechanisms of translation initiation in eukaryotes through the characterization of RNA-binding proteins (RBPs) interacting with mRNAs. Internal ribosome entry sites (IRES) are non-coding RNA regions that substitute the function of the 5’ terminal cap of mRNAs, the anchoring point of the translation machinery. Our studies allowed the identification of proteins modulating IRES activity and the understanding of structural constraints which are essential for IRES activity. We have shown that Gemin5 interacts with viral IRES as well as a selective group of cellular mRNAs. Gemin5 is a multitasking protein that forms part of the survival of motor neuron (SMN) complex, and also performs a translation regulatory role. The N-terminal domain of Gemin5 is involved in the interaction with the ribosome and the snRNAs, whereas the C-terminal region harbors a dimerization module and a non-conventional RNA-binding site (RBS). The dimerization domain consisting of a tetratricopeptide (TPR)-like domain that self-assembles into a canoe-shaped dimer.
Disruption of the dimerization capacity of Gemin5 abrogates translation enhancement induced by the C-terminal fragment of the protein. The RBS moiety harbors an intrinsically disordered region (IDR) which coevolved with the RNA-interacting region, revealing the evolutionary selection of the RNA-protein interaction module. Identification of the RNA partners of the RBS domain unveiled a feedback loop with its own mRNA, counteracting the negative effect of Gemin5 on translation. Notably, Gemin5 variants recently found in patients with neurological disorders harbor structural defects that impair dimerization, reduce translation, and show strong protein interactome decrease. In summary, this protein provides an important platform for RNP interactions, controlling translation of selective mRNAs, including ribosomal protein and histones, critical for cell growth, beyond being a key actor in spliceosome assembly.
Miembros del grupo

Encarnación Martínez Salas
Lab.: 309 Ext.: 4619
emartinez(at)cbm.csic.es

Jorge Ramajo Alonso
Lab.: 309 Ext.: 4649
j.ramajo(at)csic.es

Salvador Abellán Pérez
Lab.: 309 Ext.: 4649
sabellan(at)cbm.csic.es

Azadeh Nahavandi Araghi
Lab.: 309 Ext.: 4619
Publicaciones representativas

Structural basis for the dimerization of Gemin5 and its role in protein recruitment and translation control
María Moreno-Morcillo et al.
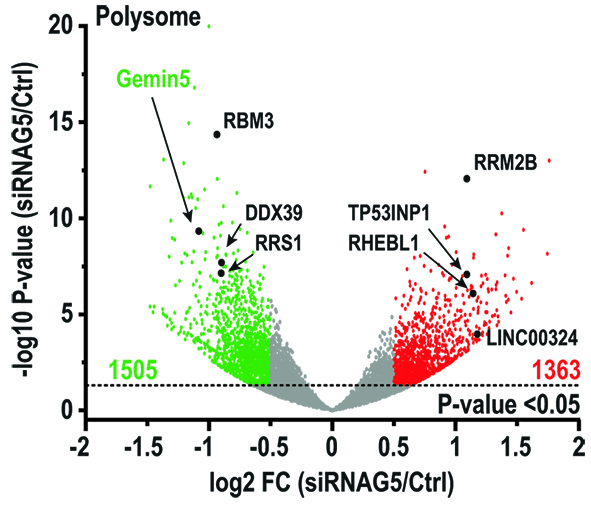
Gemin5-dependent RNA association with polysomes enables selective translation of ribosomal and histone mRNAs
Azman Embarc-Buh et al.
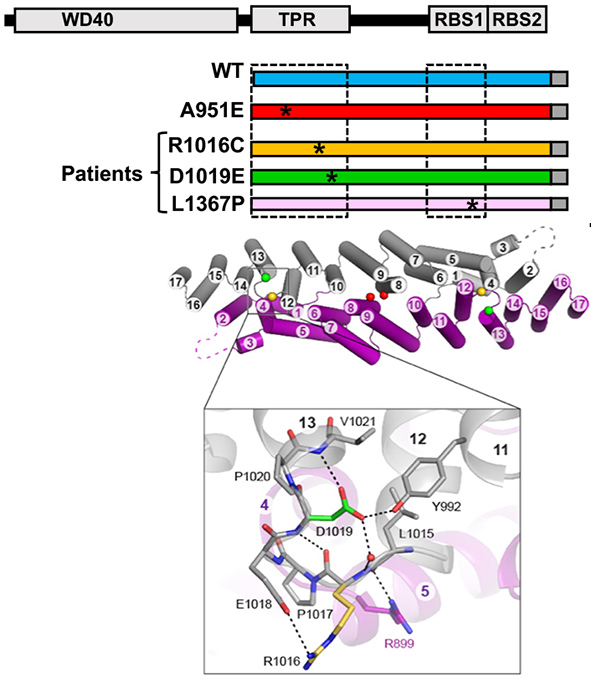
Functional and structural deficiencies of Gemin5 variants associated with neurological disorders
Rosario Francisco-Velilla et al.

Structural basis for Gemin5 decamer-mediated mRNA binding
Qiong Guo et al.