Scientific Program
Genome dynamics and function
RESEARCH GROUP
Genetics and cell biology of cancer: T-cell lymphoblastic neoplasms
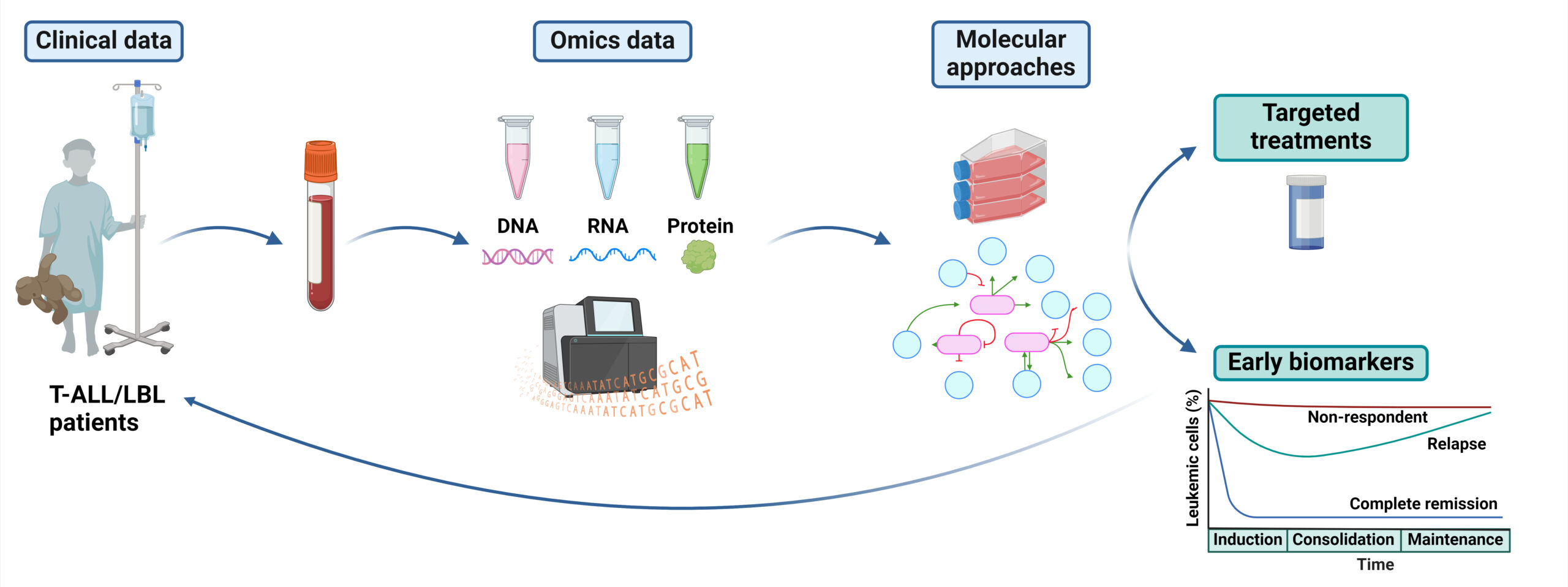
How to improve the clinical management of T-ALL/LBL patients? Using omics and clinical patient information and molecular approaches, we aim to 1) identify early biomarkers that can predict the response to therapy, allowing to refine the therapeutic stratification, and 2) identify therapeutic targets that are amenable to pharmacological intervention and offer alternative and efficient treatments.
Research
T-cell lymphoblastic neoplasms (T-ALL/LBL) are a type of aggressive hematological cancer derived from immature T-cells, mostly affecting children and adolescent boys. Current treatments consist of high-dose multi-agent chemotherapy followed by hematopoietic stem cell transplantation in standard-high risk patients. Cure rates reach up to 90% among pediatric patients but only 48% in adults, and they are associated with acute and long-term toxicity. In addition, 20-40% of patients experience a relapse or do not respond to first-line treatments, then showing a dismal prognosis with survival rates below 10% and very limited therapeutic options. These weaknesses in the clinical management of T-ALL/LBL patients need to be addressed. The lack of clinical features or molecular markers that can accurately allocate T-ALL/LBL patients to a specific risk stage at the time of diagnosis argues, at least in part, for the toxicity associated with the treatment.
On the other hand, no targeted treatments against the main alterations identified T-ALL/LBL are currently available in the clinical setting and since the approval of nelarabine in 2005 no pharmacological inhibitors have been approved for relapsed/refractory (R/R) T-ALL/LBL patients. All this highlights two unmet needs: 1) a more refined stratification of T-ALL/LBL patients according to their potential risk, which would lead to improved cure rates and reduced toxicity; and 2) the development of targeted treatments against the main alterations affecting T-ALL/LBL patients, which would contribute to lowering chemotherapy doses to avoid late effects and would offer a therapeutic option for R/R patients. Our group is trying to contribute to solve these two issues. Using genomic, transcriptomic and clinical data from patients, therapy prioritization algorithms based on the patient’s omics characteristics, together with molecular approaches to validate our hypotheses, we explore the genetic and molecular alterations in patients to identify molecular markers that are predictive of poor response to treatment and that can be obtained at diagnosis. In addition, we aim to identify potential therapeutic targets amenable to pharmacological intervention. As members of Fundación Jiménez Díaz Health Research Institute, our research involves the active participation of our colleagues from the Haematology and Pathologic Anatomy services. For years, we have been working closely together to assign a clinical significance to our molecular results. With our research, we expect to contribute to a better clinical management of this disease, with a better identification of high-risk patients to guide their treatment, and with an enhanced efficacy of current therapies in a relevant fraction of patients whose targeted treatment is nowadays an unmet need. Ultimately, this would contribute to increase the cure rates for T-ALL/LBL patients.
Group members

Javier Santos Hernández
Lab.: 409/415.4 Ext.: 4627
jsantos(at)cbm.csic.es

María Consuelo Villa Morales
Lab.: 409/415.4 Ext.: 4627
mvilla(at)cbm.csic.es

Concepción Vaquero Lorenzo
Lab.: 409 Ext.: 4653
cvaquero(at)cbm.csic.es

Laura Vela Martín
Lab.: 409 Ext.: 4653
lvela(at)cbm.csic.es

Sara Ruiz García
Lab.: 409 Ext.: 4653
s.garcia(at)cbm.csic.es
Selected publications
Comprehensive characterization of a novel, oncogenic and targetable SEPTIN6::ABL2 fusion in T-ALL
Antonio Lahera et al.
SOCS3 deregulation contributes to aberrant activation of the JAK/STAT pathway in precursor T-cell neoplasms
Antonio Lahera et al.
Quantifying telomeric lncRNAs using PNA-labelled RNA-Flow FISH (RNA-Flow)
Iria González-Vasconcellos et al.
Patterns of Differentially Expressed circRNAs in Human Thymocytes
Pilar López-Nieva et al.