Scientific Program
Genome dynamics and function
RESEARCH GROUP
Chromosome replication and genome stability

José Antonio Tercero
Our aim is to contribute to the understanding of how cells avoid genomic instability, a hallmark of cancer and other diseases. We mainly investigate the mechanisms that help maintain genome stability during chromosome replication, especially in the presence of DNA damage or replicative stress, using Saccharomyces cerevisiae as a eukaryotic model.
Research
The maintenance of genome integrity during chromosome replication and the fidelity of DNA synthesis are essential for the correct transmission of genetic information in each cell division cycle. Inevitably, chromosome replication is threatened by DNA damage and replicative stress, which constitute a potential source of errors and a risk for the stability and progression of replication forks. Successful genome duplication in each cell cycle requires repair or tolerance of DNA lesions, protection of replication forks, and the ability to resume DNA synthesis after fork stalling. Failures in these processes lead to genomic instability, a hallmark of cancer cells as well as a major cause of premature ageing and some developmental and neurological disorders.
The aim of our group is to contribute to the knowledge of the mechanisms that eukaryotic cells use to maintain genome stability during chromosome replication, especially under conditions of DNA damage or replicative stress. We study how DNA repair, DNA damage tolerance and checkpoint pathways, in conjunction with different proteins such as helicases and nucleases, facilitate chromosome replication in the presence of DNA lesions or replication perturbations. We analyse the contribution of these proteins to the integrity and function of DNA replication forks, their regulation and their importance for cell viability and the prevention of genomic instability. The main aspects of these processes are evolutionarily conserved, which allows us to study them in detail using the yeast Saccharomyces cerevisiae as a working model organism.
Group members

José Antonio Tercero Orduña
Lab.: 404 Ext.: 4818
jatercero(at)cbm.csic.es

Raquel Plaza Trujillo
Lab.: 404 Ext.: 4818
rplaza(at)cbm.csic.es

Karolina Mikulska
Lab.: 404 Ext.: 4516
Selected publications
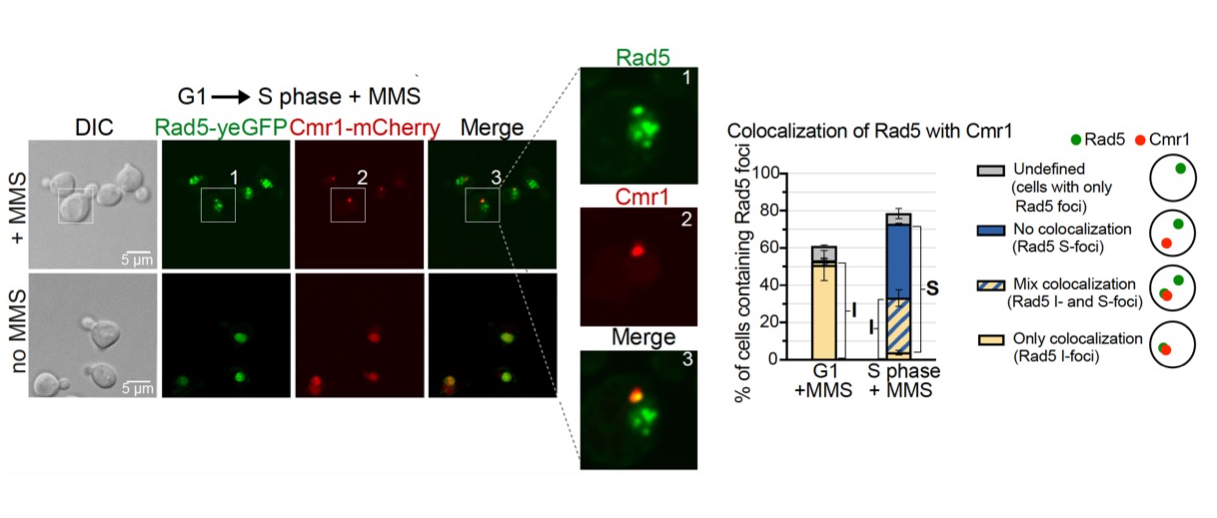
Spatial regulation of DNA damage tolerance protein Rad5 interconnects genome stability maintenance and proteostasis networks
Carl P Lehmann et al.
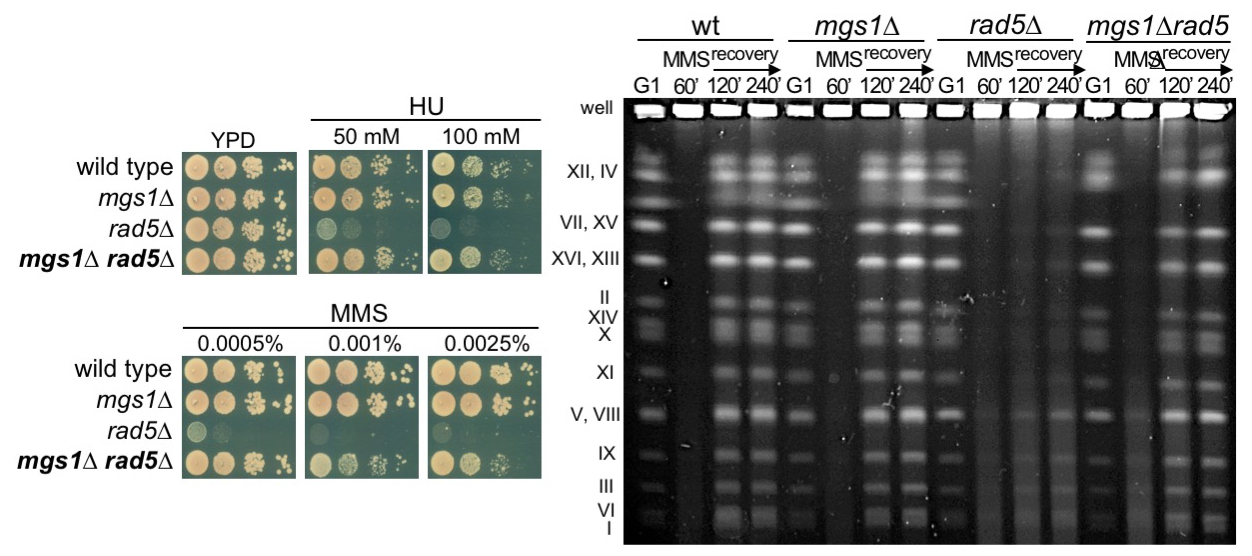
The Mgs1/WRNIP1 ATPase is required to prevent a recombination salvage pathway at damaged replication forks
Alberto Jiménez-Martín et al.
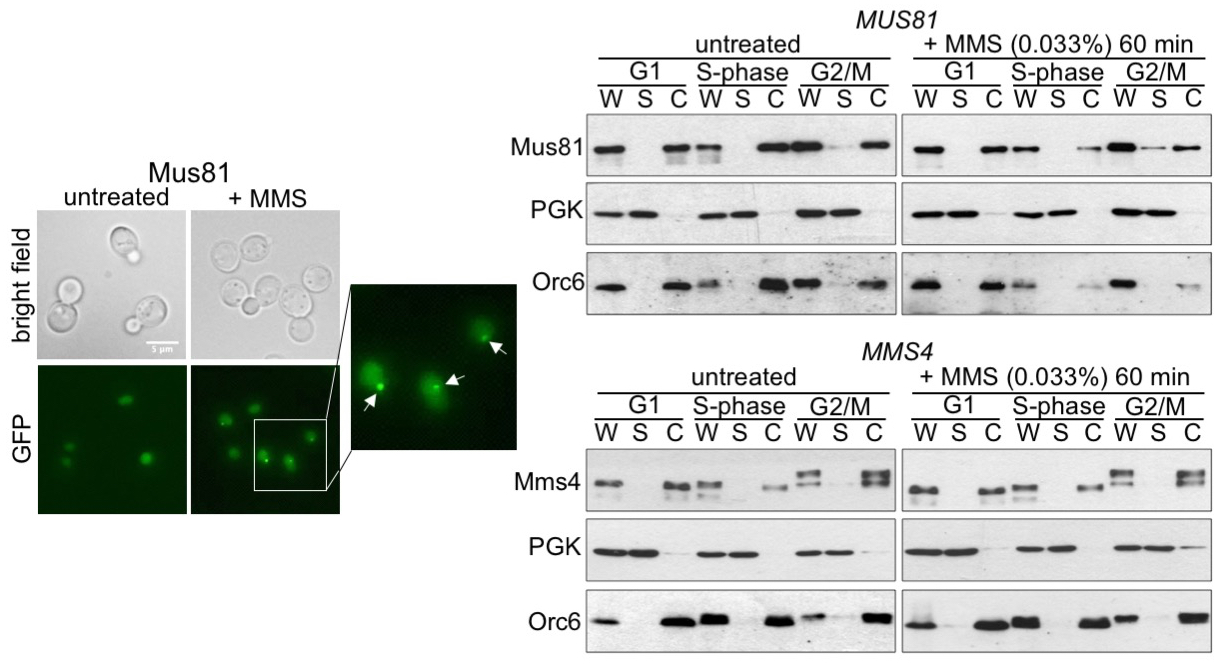
Subnuclear Relocalization of Structure-Specific Endonucleases in Response to DNA Damage
Irene Saugar et al.
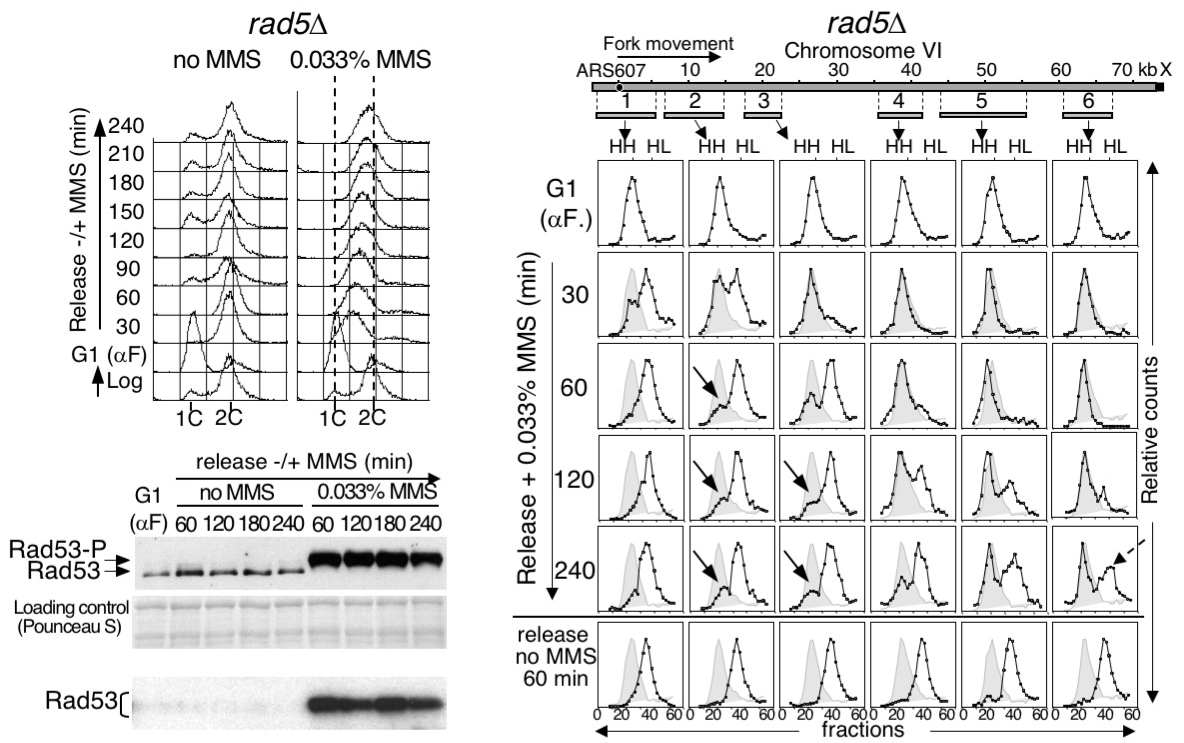
Rad5 Plays a Major Role in the Cellular Response to DNA Damage during Chromosome Replication
María Ángeles Ortiz-Bazán et al.