Scientific Program
Interactions with the environment
RESEARCH GROUP
Modulation of antiviral immunity by viral proteases and noncoding RNAs

Margarita Sáiz Zalabardo
We are interested in deciphering the molecular processes evolved by RNA viruses to counteract the host antiviral response, particularly those involving viral proteases. The interplay between viral genomes and proteins with cellular sensors is crucial for the activation of an effective immune response and may determine the outcome of disease. We also work in the characterization of viral non-coding RNA regions and their use as antiviral strategies.
Research
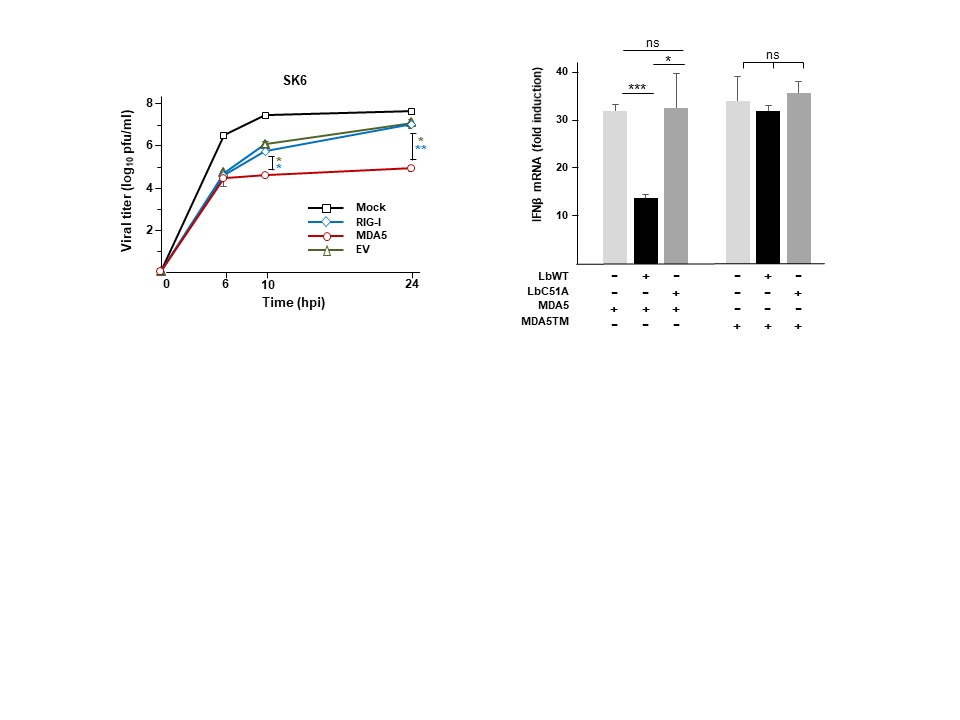
Our research interest is focused on the study of the interactions between RNA viruses and host cells involving the non-coding regions (NCR) of the viral genomes and the catalytic activity of the viral proteases. Our studies include the identification of proteins sensing infection and viral RNA genomes, as well as the characterization of the immune evasion molecular mechanisms exerted by the virus to counteract the host antiviral response based on triggering of the type-I interferon pathway in infected cells. The outcome of the balance between the antiviral response and viral antagonism may determine the onset of disease and pathogenesis. We use foot-and-mouth disease virus (FMDV), a highly infectious and worldwide distributed pathogen, as a model and we are actively working on the identification of host innate effector proteins which are targets for the proteolytic activity of the two virally encoded proteases (Lpro and 3Cpro) for interference with different signaling routes known to respond to infection by RNA viruses. We have reported a novel mechanism of viral evasion based on the dual cleavage of LGP2 and MDA5, immune sensors of the RIG-I-like receptors family, at a conserved helicase motif by the FMDV Leader protease (Lpro) and the redundant degradation of cGAS by 3Cpro and Lpro.
These interactions have a negative impact on antiviral immunity promoting, in turn, viral infection. We also work in the characterization of the structural domains in the FMDV 5´ and 3´NCR of the FMDV genome, including their RNA-RNA, RNA-protein interactions and structural requirements involved in virus viability, infectious capacity and innate immune response elicited. Our previous studies on the FMDV NCRs have led us to test the biotherapeutic applications of synthetic non-coding RNAs derived from the FMDV 5´and 3´NCR (ncRNAs) and we have shown their ability to induce a broad spectrum antiviral activity and to enhance specific B- and T-cell mediated immune responses elicited after vaccination. We have reported the inhibitory effect of the ncRNAs against infection by pathogens of different viral families including relevant zoonotic viruses (FMDV, West Nile virus, Rift Valley fever virus, African swine fever virus) and shown their biological activity in cells from farm and wild animals, as well as in vivo in mice and swine. The pan-coronaviruses antiviral activity of these ncRNAs has been recently addressed showing the intrinsic prophylactic activity of naked RNA against SARS-CoV-2 in a COVID-19 mouse model. We are also working on the development of new nanotechnology-based formulations to improve the ncRNAs delivery. The results derived from these studies will contribute to gain knowledge on the interaction between viral pathogens and the innate immune system of the host cell and to design new and more effective strategies against current and newly emerging viruses. Also, learning from how viruses counteract host immune responses through specific targeting of relevant effector proteins may help in the design of new therapeutic strategies against type I interferonopathies.
Group members

Margarita Sáiz Zalabardo
Lab.: 105 Ext.: 4729
msaiz(at)cbm.csic.es

Miguel Ramón Rodríguez Pulido
Lab.: 105 Ext.: 4522
mrrodriguez(at)cbm.csic.es

Catalina Grabowski Pinto
Lab.: 105 Ext.: 4522
cgrabowski(at)cbm.csic.es

Miryam Polo Hernández
Lab.: 105 Ext.: 4522
mpolo(at)cbm.csic.es
Selected publications
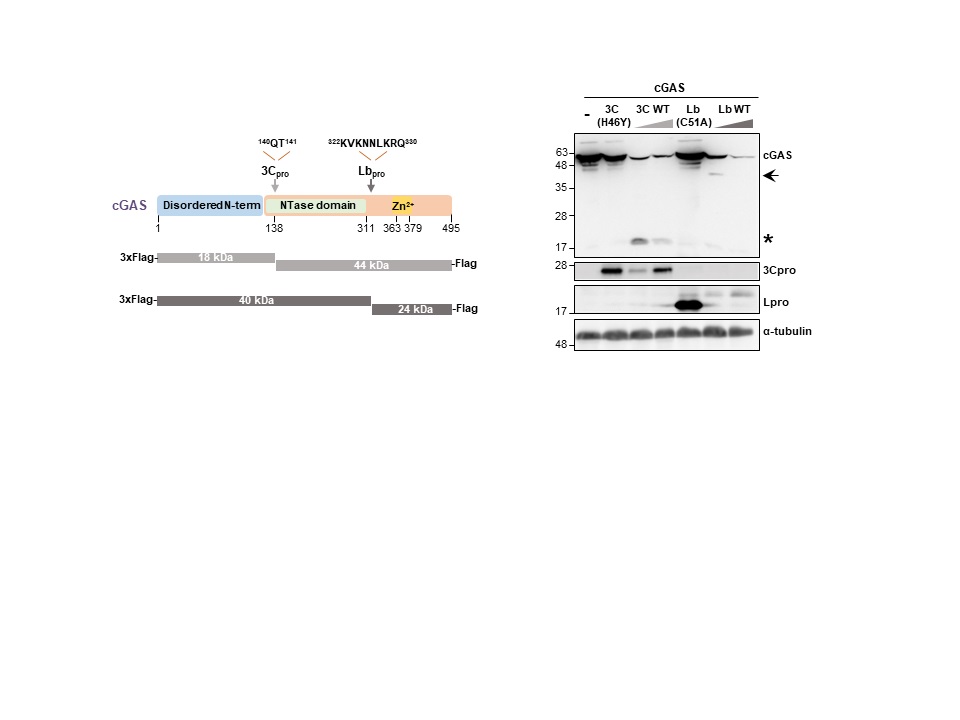
The antiviral response triggered by the cGAS/STING pathway is subverted by the foot-and-mouth disease virus proteases
Miguel Ángel Sanz et al.

MDA5 cleavage by the Leader protease of foot-and-mouth disease virus reveals its pleiotropic effect against the host antiviral response
Miguel Rodríguez Pulido et al.
Non-coding RNAs derived from the foot-and-mouth disease virus genome trigger broad antiviral activity against coronaviruses
Miguel Rodriguez Pulido et al.
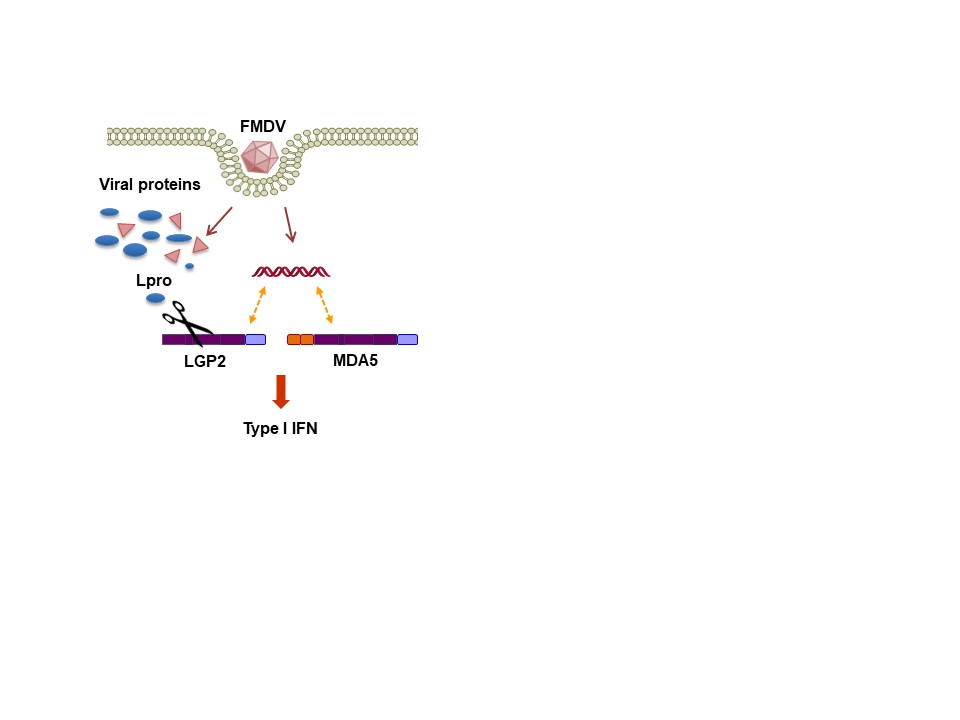
Innate immune sensor LGP2 is cleaved by the Leader protease of foot-and-mouth disease virus
Miguel Rodríguez Pulido et al.