Scientific Program
Interactions with the environment
RESEARCH GROUP
Ultrahigh-throughput discovery and engineering of enzymes for biotechnological applications

Aurelio Hidalgo Huertas
Nature’s vast genetic reservoir constitutes an unfathomable wealth of biogenics for healthier, cleaner, more sustainable living. In the HT Discovery Lab, we develop and use state-of-the-art screenings to uncover novel biogenics for the next generation of fair, sustainable and effective pharma, industrial and consumer products, supporting the development of a European circular bioeconomy
Research
Microbial diversity is a vast reservoir of genetic information that can be valorized through industrial application, from biosynthetic gene clusters to novel protein catalysts. The synergy between new experimental discovery tools based on biology and those based on nanotechnologies are instrumental to find relevant genes faster and more efficiently, enabling especially academic labs to undertake screening campaigns until now costly and limited to large enterprises.
In our laboratory, we make use of both biological selections or ultrahigh-throughput screening based on droplet microfluidics to find rare genes and enzymes of interest in the natural or man-made diversity. Biological selections couple the improved fitness of a protein to the survival of a biological host under selective pressure. Using these and other methods for protein engineering, we have developed “thermostable” and soluble variants of enzymes for biocatalysis, such as esterases, dehalogenases and aldolases and fluorescent proteins.
However, the complexity of cellular metabolism limits the application of biological selections. Microfluidics enables the miniaturization of assays with throughput of kHz and a 1000x reduction in volume and costs. Therefore, droplet microfluidics achieves throughputs typical of biological selections with none of their complexity. Within the H2020 project MetaFluidics, in our laboratory, we have implemented a fluorescence-based microfluidic sorting platform and assays to find stable hydrolases in thermal environments as well as other relevant enzymes for biocatalysis. This has entailed developing news hosts and methods for functional expression, compatible with enzymatic assays at high temperatures.
Group members

Patricia Pérez Arnaiz
Lab.: 108 Ext.: 4525
patricia.perez(at)cbm.csic.es

Aurelio Hidalgo Huertas
Lab.: 108 Ext.: 4527
ahidalgo(at)cbm.csic.es

Jorge Diaz-Rullo Aroco
Lab.: 108 Ext.: 4524
jdiaz(at)cbm.csic.es

Davide Agostino Cecchini
Lab.: 108 Ext.: 4525
davide.cecchini(at)uam.es

Nargisse Nejda Hamdoun
Lab.: 108 Ext.: 4524
n.nejda(at)cbm.csic.es

Laura Blas Muñoz
Lab.: 108 Ext.: 4525
laura.blas(at)cbm.csic.es

Laura Pilar Saiz Alvarez
Lab.: 108 Ext.: 4525
laura.saiz(at)cbm.csic.es

Gonzalo Rodrigo Albert
Lab.: 108 Ext.: 4525
gonzalo.rodrigo(at)cbm.csic.es

Sara Trujillo Cubillo
Lab.: 108 Ext.: 4525

Pablo Rodríguez Mateos
Lab.: 108 Ext.: 4527
prodriguez(at)cbm.csic.es
Selected publications
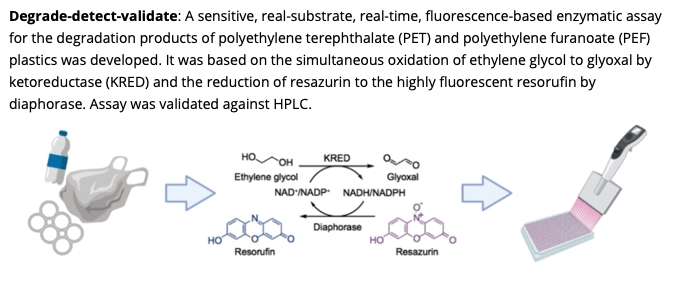
A Coupled Ketoreductase-Diaphorase Assay for the Detection of Polyethylene Terephthalate-Hydrolyzing Activity
Dr. María Gimeno-Pérez et al.
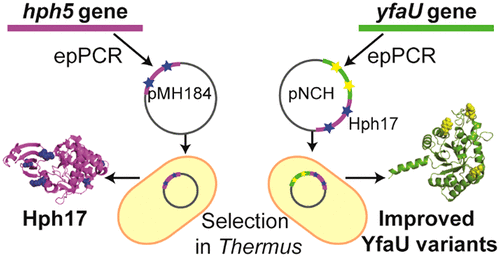
Thermostability Engineering of a Class II Pyruvate Aldolase from Escherichia coli by in Vivo Folding Interference
Sandra Bosch et al.
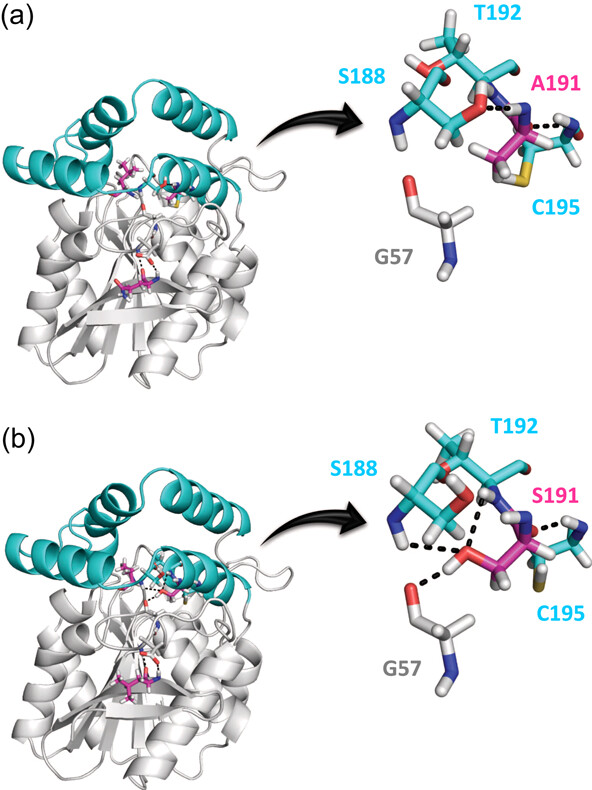
Thermostability enhancement of the Pseudomonas fluorescens esterase I by in vivo folding selection in Thermus thermophilus
Diana M. Mate et al.
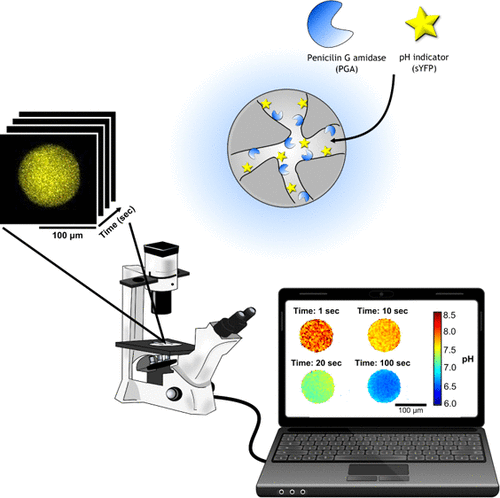
Biobased, Internally pH-Sensitive Materials: Immobilized Yellow Fluorescent Protein as an Optical Sensor for Spatiotemporal Mapping of pH Inside Porous Matrices
Tanja Consolati et al.