Scientific Program
Genome dynamics and function
RESEARCH GROUP
Maintenance of bacterial genome stability

Miguel de Vega José
To understand how the faithful and efficient transmission of genetic information is carried out, we study the biochemical properties of DNA polymerases that enable the enzyme to efficiently and accurately replicate and repair the genome. For this purpose, we perform biochemical assays with purified DNA polymerases and analyze their behavior on different DNA substrates.
Research
Maintenance of genome stability largely relies on faithful DNA replication. However, the continuous damage of the genomes by genotoxic agents has rendered necessary the emergence of repair mechanisms to prevent the deleterious effects that permanence of such damages could cause.
Our main objective is to get insights on the molecular mechanisms responsible for maintaining genome stability in bacteria, by functional analysis of the enzymatic features of purified repair proteins from model organisms as the gram positive bacterium Bacillus subtilis whose vegetative cells and spores have to handle DNA damage induced by extreme environmental conditions, and the gram negative Pseudomonas aeruginosa.
In this sense, during the last years we have been studying the catalytic functions of the B. subtilis DNA polymerase belonging to family X (PolXBs). We have shown that, in addition to the polymerization activity, PolXBs possesses an intrinsic 3’-5’ exonuclease, AP endonuclease, 3’-phosphatase and 3’-phosphodiesterase activities that share a common catalytic core at the C-terminal PHP domain, specifically present in the bacterial/archaeal subgroup of PolXs. Acting in concert with polymerization, those activities endow bacterial PolXs with the faculty to perform abasic site (AP) recognition, incision and further restoration (repair) of the original non-damaged nucleotide, as well as processing of the 3’-damaged ends that can arise after the exposition of the DNA to genotoxic agents.
Many bacterial members are provided with a nonhomologous end joining system (NHEJ) responsible for repairing double strand breaks (DSB), the most hazardous DNA lesions as they are lethal to dividing cells if they are not repaired in a timely fashion. Such a repair pathway is constituted by a Ku homodimer and a dedicated and multifunctional ATP-dependent Ligase (LigD). Previous biochemical analysis of bacterial LigD allowed the identification of polymerization, ligase and fosfoesterase activities. We have recently characterized the additional presence of an unexpected 5’-2-deoxyribose-5-phosphate (dRP) lyase activity in the ligase domain (LigDom) of the LigD from B. subtilis and P.aeruginosa. This activity coordinates with the polymerization and ligase activities to allow efficient repairing of an AP site-containing DNA in an in vitro reconstituted Base Excision Repair (BER) reaction. Therefore, LigD has in the same polypeptidic chain the three activities required in the last steps of BER, suggesting that its DNA repair role is not restricted to the NHEJ pathway, but expands beyond, being potentially active in additional repair pathways. Additionaly we have identified a novel AP-lyase activity in the PolDom of LigD that cleaves AP sites specifically when they are proximal to recessive 5’-ends and through the formation of a preternary precatalytic complex with Mn2+ ions and an incoming ribonucleotide complementary to the templating nucleotide opposite the AP site, guaranteeing the coupling of AP sites removal to the end-joining reaction by the bacterial LigD.
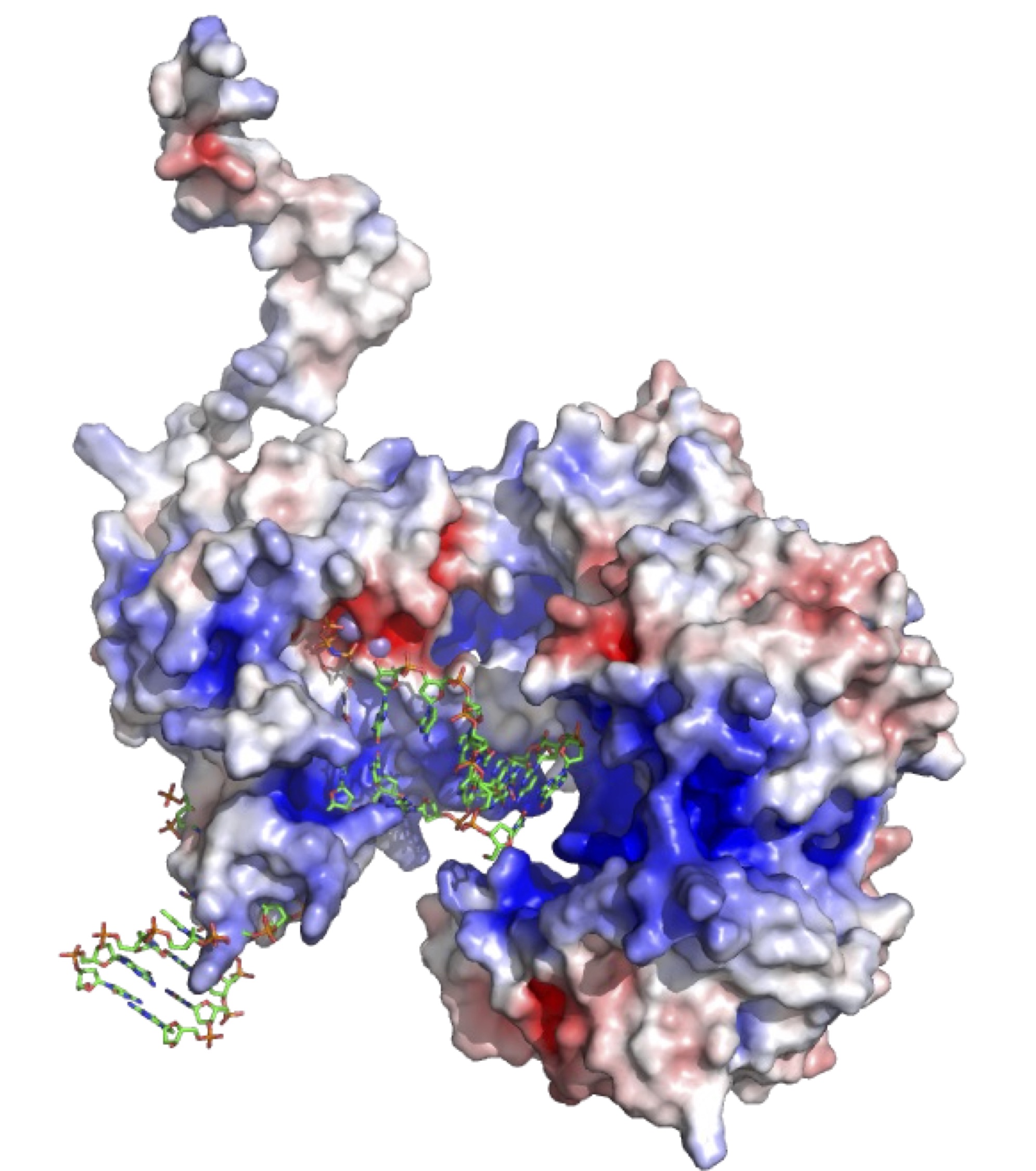
We have recently characterized a protein called Insertion Sequence excision enhancer, specifically present in enteropathogenic E. coli bacteria, and belonging to the Archaeal/Eukaryotic primase superfamily. Our studies have demonstrated that this is a novel DNA polymerase capable of carrying out microhomology-mediated end joining, possessing biochemical characteristics typical of DNA polymerases involved in DNA lesion repair.
Group members

Miguel de Vega José
Lab.: 405 Ext.: 4717
mdevega(at)cbm.csic.es

Alicia del Prado Díaz
Lab.: 405 Ext.: 4463
adelprado(at)cbm.csic.es

Silvia Díaz Arco
Lab.: 405 Ext.: 4463
silvia.diaz(at)cbm.csic.es

Lucía Alba Fernández
Lab.: 405 Ext.: 4463
lucia.alba(at)cbm.csic.es

Aitana García García
Lab.: 405 Ext.: 4463
Selected publications
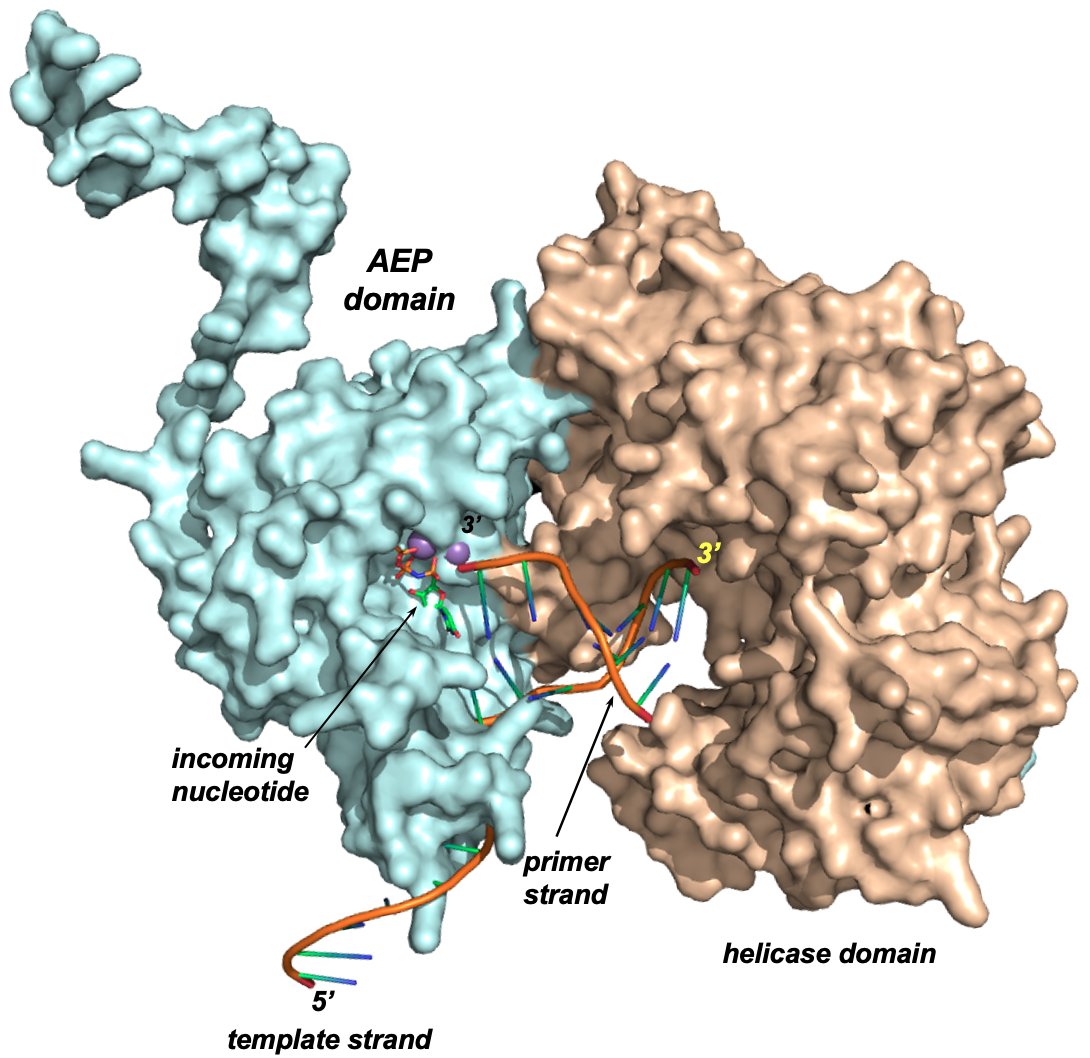
The enterohemorrhagic Escherichia coli insertion
sequence-excision enhancer protein is a DNA
polymerase with microhomology-mediated
end-joining activity
Patricia A. Calvo et al.
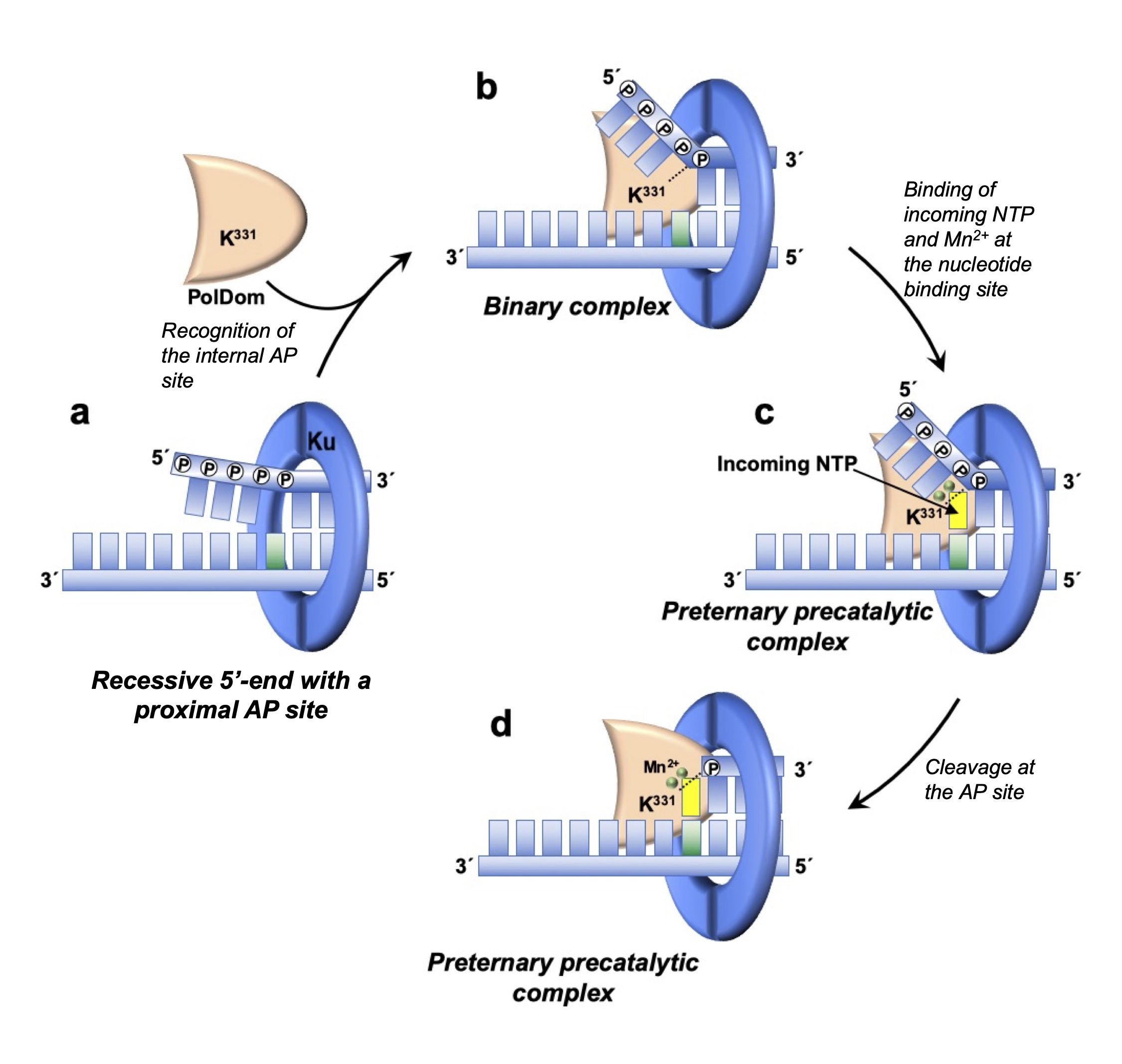
Bacterial Ligase D preternary-precatalytic complex performs efficient abasic sites processing at double strand breaks during nonhomologous end joining
Ana de Ory et al.
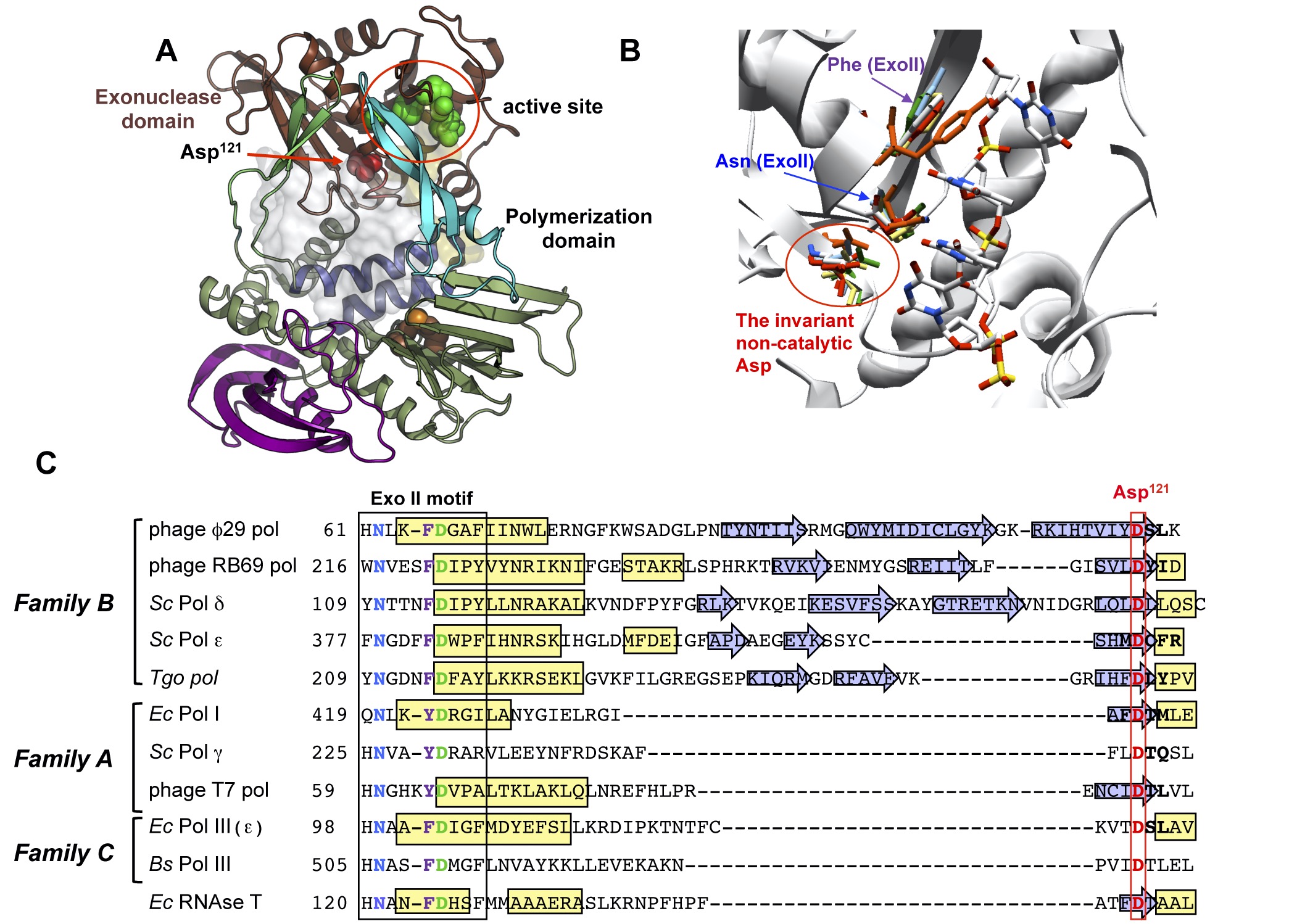
Noncatalytic aspartate at the exonuclease domain of proofreading DNA polymerases regulates both degradative and synthetic activities
Alicia del Prado et al.
Efficient processing of abasic sites by bacterial nonhomologous end-joining Ku proteins
Ana de Ory et al.