Scientific Program
Genome dynamics and function
RESEARCH GROUP
Regulation of mRNA translation in eukaryotes and its implications for organismal life
Translation control in eukaryotes and its role in cellular homeostasis
We investigate how eukaryotic systems regulate translation, both at global and message-specific manner. Our goal is to identify new regulatory elements in mRNAs and ribosomes and new translation initiation factor (eIF) activities involved in translation control during cell proliferation, stress response and survival in human cells and yeast models. We study how translation reprogramming through eIF2 phosphorylation in response to stress modulates proteostasis and autophagy. To do this, we combine genetic analysis with structural modeling, real time quantification of gene expression and genome wide analysis of translation by polysome profiling and RNAseq. We also study how some viruses interfere with host translation by partially blocking ribosomal function, which is a powerful work tool.

Research
We have identified the ES6S region of 40S ribosomal subunit as the gateway for mRNA entry and 43S-PIC scanning during translation initiation. Our data suggest the ES6S region could be serving as a platform for the recruitment of RNA helicases (eIF4A and DDX3, among others) involved in RNA secondary structure unwinding. Blocking ES6S region differentially affected translation of mRNAs encoding some proto-oncogenes (H-Ras, CCND3, ODC-1, etc.), genes involved in signal transduction with long and structured 5´UTR mRNAs and also some viral mRNAs. We are currently evaluating the ES6S region as a target for antitumoral and antiviral molecules.
More recently, we found that the differential effect of the non-structural protein 1 (NSP1) of SARS-CoV-2 on translation depends on the composition of 5’ UTR and codon usage bias of target mRNA. Our data suggest the existence of two mechanisms for the recruitment of mRNA to the 40S subunit (threading and slotting) and a functional interaction between the speed of elongation (codon bias) and translation initiation. We are also looking at the implications of our results for the molecular evolution of SARS-CoV-2 and other human respiratory viruses.
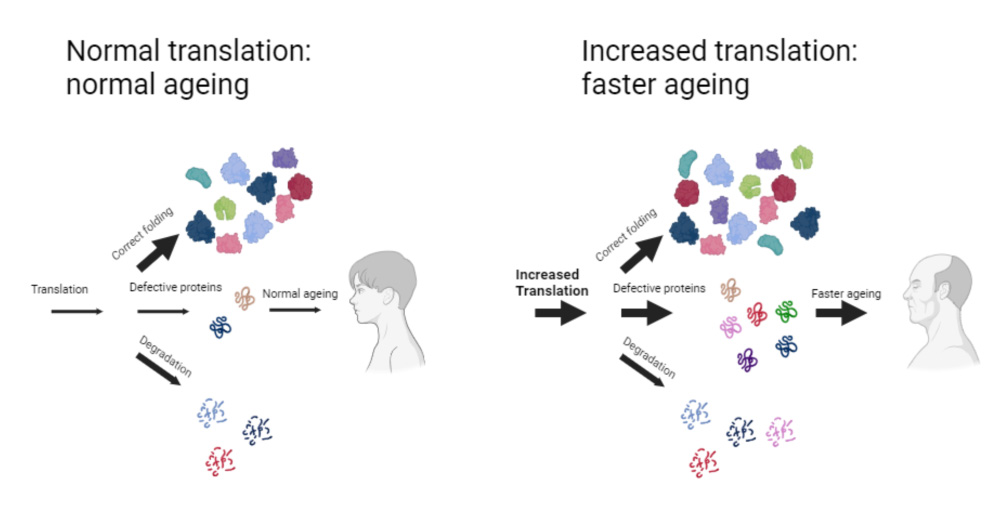
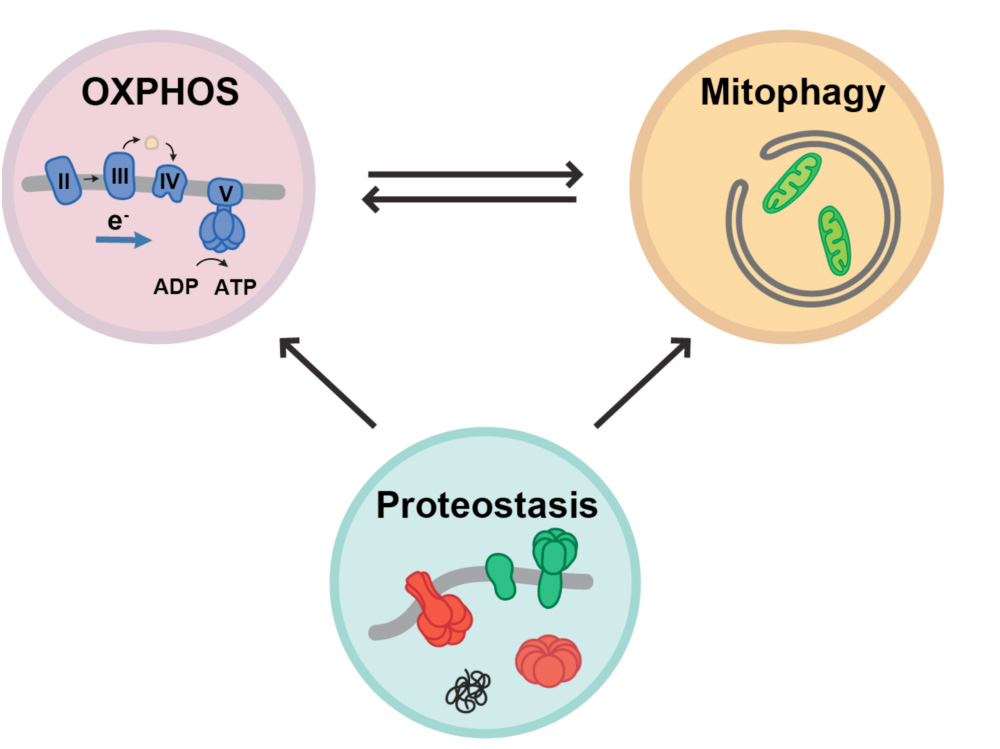
We continue to study the molecular and functional links among stress response, translational reprogramming and cellular homeostasis. Thus, we found that preventing eIF2α phosphorylation in yeast not only impaired stress response, but also accelerated aging by a mechanism that involves proteostasis and autophagy disruption. We have identified some genes involved in proteasome activity whose expression is regulated by eIF2α phosphorylation. We are characterizing the role of these genes in key processes regulating proteostasis such as protein aggregation, autophagy, mitophagy and how they can affect cell longevity.
Group members

Juan José Berlanga Chiquero
Lab.: 106 Ext.: 4714/4534
jberlanga(at)cbm.csic.es

Iván Ventoso Bande
Lab.: 106/114.4( despacho) Ext.: 4809
iventoso(at)cbm.csic.es

Miguel Angel Rodríguez Gabriel
Lab.: 106 Ext.: 4816
marodriguez(at)cbm.csic.es

Margarita Cabrera Sola
Lab.: 106 Ext.: 4534
mcabrera(at)cbm.csic.es

Mercedes Núñez Bayón
Lab.: 106 Ext.: 4534
mnunnez(at)cbm.csic.es

Carla Henares Sánchez
Lab.: 106 Ext.: 4809
carla.henares(at)cbm.csic.es

Irene Rubio Serrano
Lab.: 106 Ext.: 4534
Selected publications
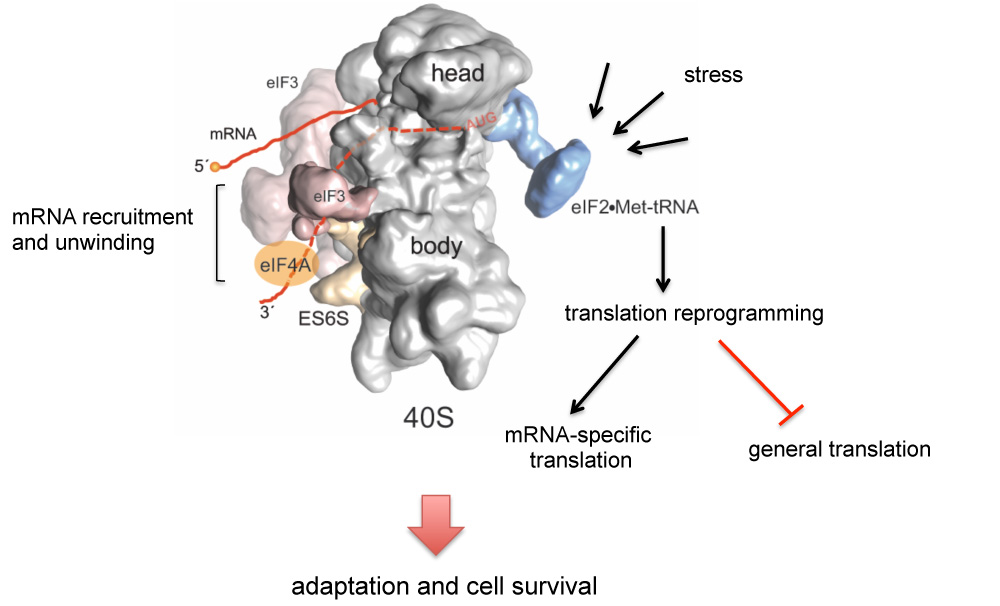
An mRNA-binding channel in the ES6S region of the translation 48S-PIC promotes RNA unwinding and scanning
Irene Díaz-López et al.
Antagonistic effects of mitochondrial matrix and intermembrane space proteases on yeast aging
Montserrat Vega et al.
Translational control of gene expression by eIF2 modulates proteostasis and extends lifespan
Tamara Jiménez-Saucedo et al.
Title
Authors





