Scientific Program
Interactions with the environment
RESEARCH GROUP
Molecular modelling

Paulino Gómez-Puertas
Computational biology lab. The work is devoted to the integration of evolutive and structural information to study the function of proteins, the simulation of dynamic processes of protein-protein and protein-ligand interaction, the development of novel “in silico” drug design systems and the generation of new quantitative methods for computational biology.

Research
Current projects:
– Analysis by computational simulation of enzymatic reactions catalyzed by enzymes of biomedical interest. Design of specific inhibitors. Cohesin: Analysis of the molecular interactions among the protein components of the cohesin ring and the interaction of the protein complex with DNA. Carbapenemases: Dynamic simulation of the interaction of bacterial carbapenemases (VIM-2, KPC-2, OXA-48) with substrates and known inhibitors.
– Development of a new and efficient drug design system based on computational dynamic simulation of macromolecular structures. Based on the analysis of enzyme active centers, the method developed by our group consists of simulating these protein structures through molecular dynamics for several hundred nanoseconds, selecting different representative structures and filtering a database of 3D compounds for each one of them. Used for the design of new carbapenemases inhibitors and the design of specific inhibitors as antitumor drugs.
– Development of efficient methods for calculating paths of minimum or maximum parameter values (e.g., minimum energy paths) through surfaces of any number of dimensions.
Group members

Paulino Gómez Puertas
Lab.: 313 Ext.: 4663
pagomez(at)cbm.csic.es

Iñigo Marcos Alcalde
Lab.: 313 Ext.: 4662
imarcos(at)cbm.csic.es
Selected publications
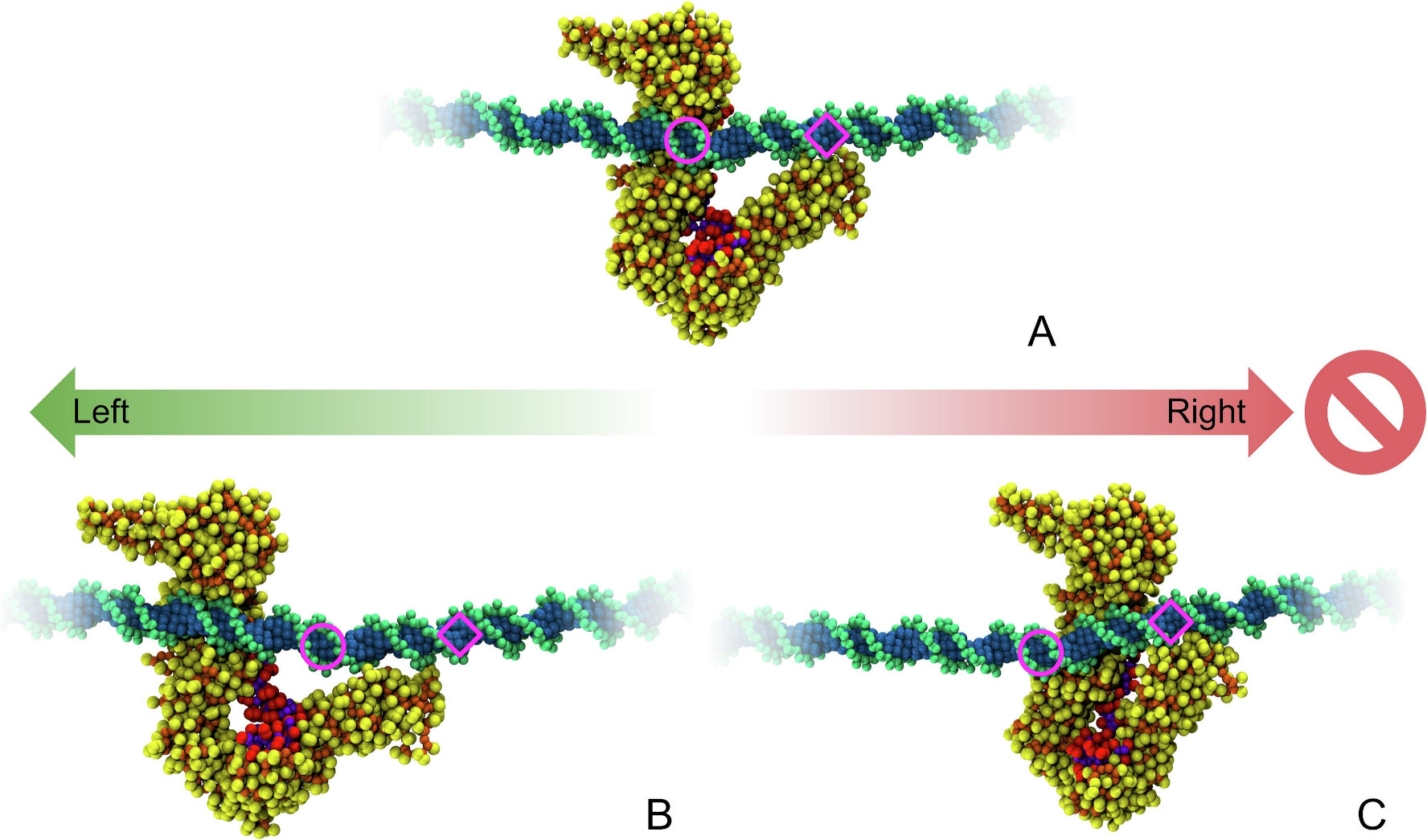
STAG2-RAD21 Complex: a Unidirectional DNA Ratchet Mechanism in Loop Extrusion
David Ros-Pardo et al.

MEPSA: minimum energy pathway analysis for energy landscapes
Iñigo Marcos-Alcalde et al.

MEPSAnd: minimum energy path surface analysis over n-dimensional surfaces
Iñigo Marcos-Alcalde et al.

STAG2: Computational Analysis of Missense Variants Involved in Disease
David Ros-Pardo et al.





