Scientific Program
Physiological and pathological processes
RESEARCH GROUP
Translational medicine in inborn errors of metabolism and other rare genetic diseases
Integrating multi-omics approaches, data analysis, experimental models, and specialized platforms offers a promising future for patients with rare genetic diseases. This intersection of diverse disciplines enables the following benefits:
• Developing precise and personalized diagnoses.
• Identifying novel therapeutic targets
We are designing advanced therapies that are more effective and safer.
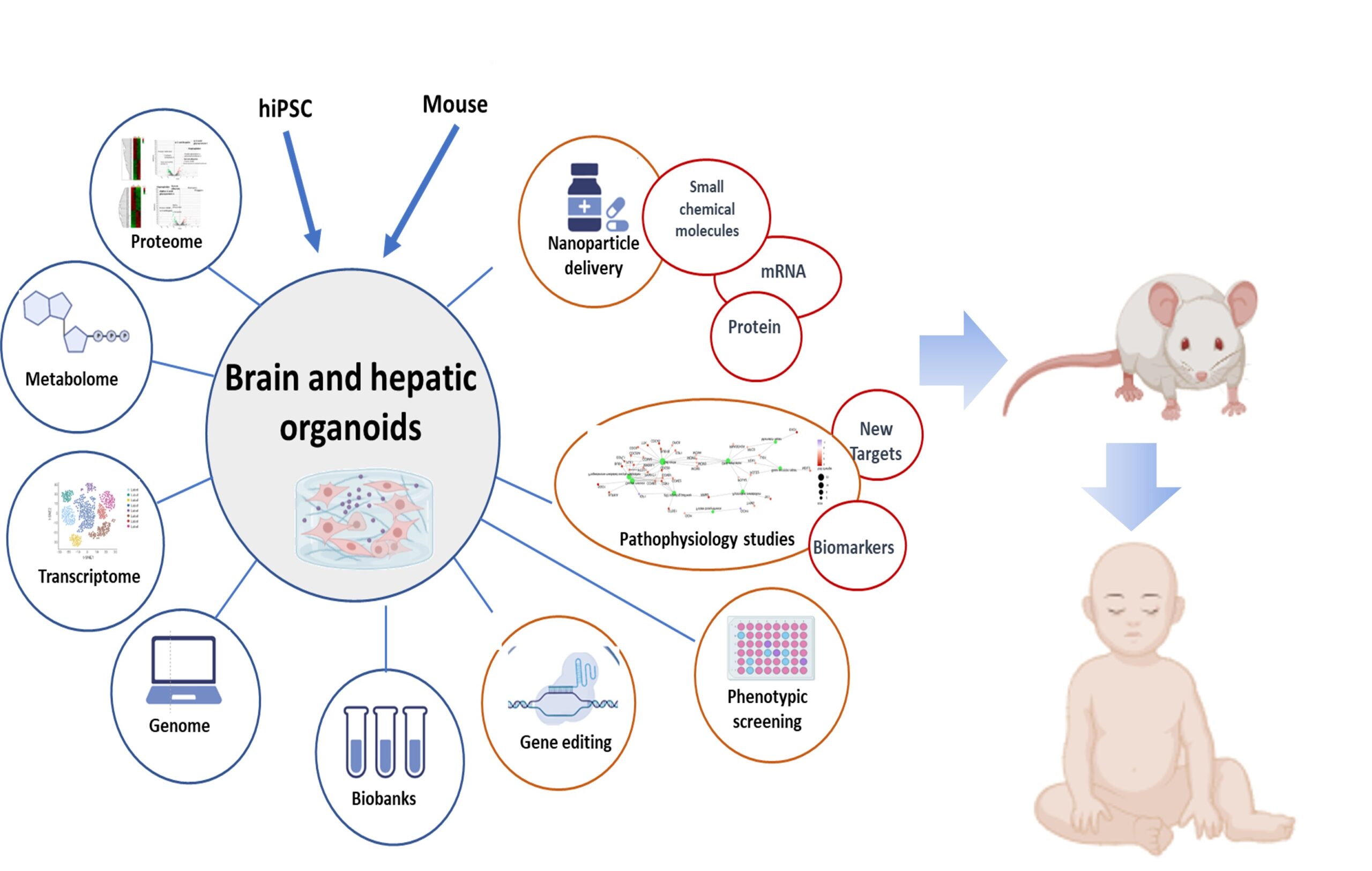
Research
Our group aims to apply the knowledge gained from basic research to clinical practice and improve the diagnosis, prevention, and treatment of inherited metabolic diseases (IMD). These diseases are prevalent, affecting 1 in every 800 newborns and belong to the largest groups of rare diseases. High-throughput genomic sequencing has helped us discover new gene-pathology associations in IEM. However, we propose a new approach to overcome the limitations of current technologies in identifying specific genetic causes of the disease. Our plan involves using third-generation sequencing, metabolomics, transcriptomics, and epigenomics to detect long repetitive elements, copy number alterations, structural variations, and epigenetic defects in DNA or RNA. We will also use a functional genomics platform to understand the clinical impact of genetic defects.
Our second objective is to research advanced therapeutic strategies using biocompatible nanoparticles loaded with small chemical compounds, repositioning drugs, RNA therapy, or therapeutic proteins. We are developing preclinical liver and brain organoid platforms obtained through human iPSC differentiation to evaluate potential drugs. We will edit, activate, or inhibit genes using CRISPR to generate models and to explore this techology as potential therapy.
Finally, we aim to identify new therapeutic targets and biomarkers by integrating multi-omics data into computational models.
Proyecto Financiado por el ISCIII (PI22/00699) y proyectos ACCI CIBERER
Group members

Belén Pérez González
Lab.: 220 Ext.: 4566/7830
bperez(at)cbm.csic.es

Pilar Rodríguez Pombo
Lab.: 220 Ext.: 4628
mprodriguez(at)cbm.csic.es

Rosa María Navarrete López de Soria
Lab.: 220 Ext.: 4596/7830
rnavarrete(at)cbm.csic.es

Mª Fátima Leal Pérez
Lab.: 220 Ext.: 4596/7830
fleal(at)cbm.csic.es

Mª Alejandra Gámez Abascal
Lab.: 220 Ext.: 4596/7830
agamez(at)cbm.csic.es

Arturo Martín Martínez
Lab.: 220 Ext.: 4596

Patricia Pascual Vinagre
Lab.: 220 Ext.: 4596

Rafael Hencke Tresbach
Lab.: 220 Ext.: 4596

Francisca Diaz González
Lab.: 220 Ext.: 4566
fdiaz(at)cbm.csic.es

Cristina Barrigas Viñuela
Lab.: 220 Ext.: 4566

María Aránzazu Jacomé Feliú
Lab.: 220 Ext.: 4666
mjacome(at)cbm.csic.es
Selected publications

Pathogenic variants of the coenzyme A biosynthesis-associated enzyme phosphopantothenoylcysteine decarboxylase cause autosomal-recessive dilated cardiomyopathy
Irene Bravo-Alonso et al.
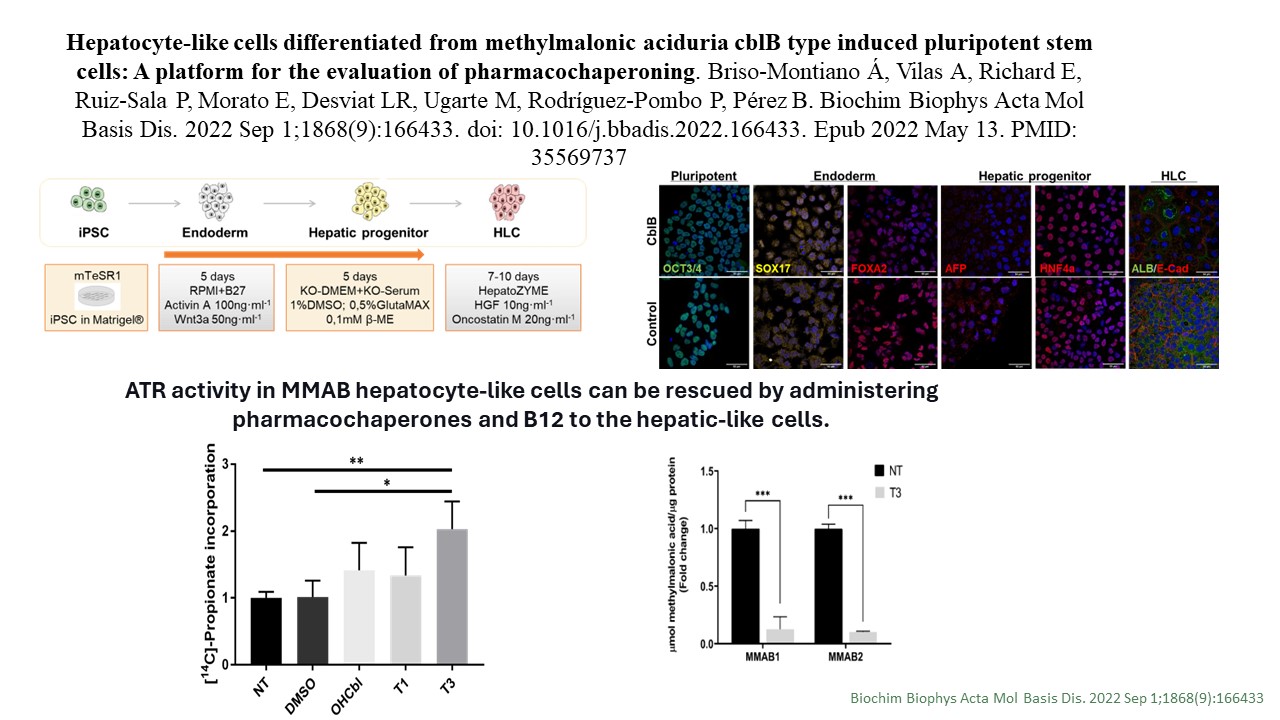
Hepatocyte-like cells differentiated from methylmalonic aciduria cblB type induced pluripotent stem cells: A platform for the evaluation of pharmacochaperoning
Á Briso-Montiano et al.
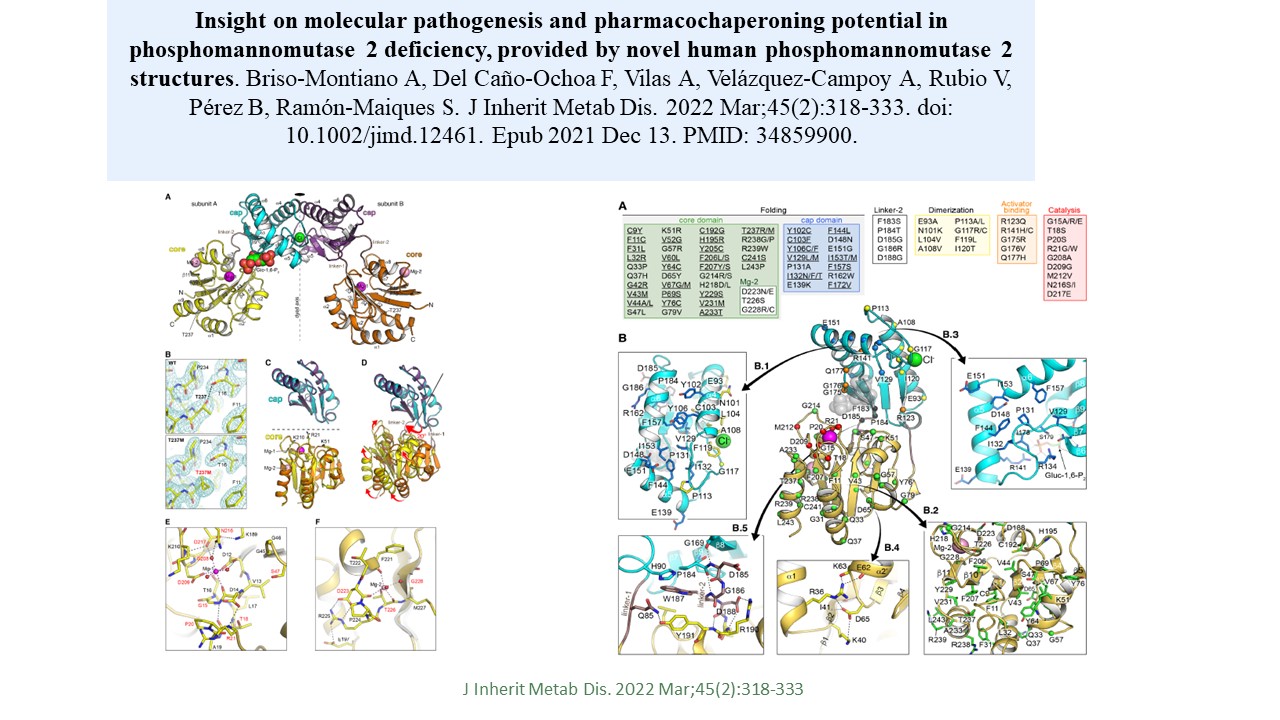
Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures
Alvaro Briso-Montiano et al.

Pathogenic variants in GCSH encoding the moonlighting H-protein cause combined nonketotic hyperglycinemia and lipoate deficiency
Laura Arribas-Carreira et al.





