DNA replication, chromatin and cell division
Research summary:
The transition to multicellularity required the acquisition of novel structures and mechanisms to coordinate cell division, acquisition of cell fates and the differentiation, and the establishment of complex regulatory networks. Our group is interested in understanding the mechanisms that control these processes and how epigenetic mechanisms affect such coordination.
To that end, we use the model plant Arabidopsis thaliana that offers us the possibility of carrying out molecular, cellular, genetic and genomic approaches. In addition, plant development, contrary to the situation in animals, is post-embryonic and occurs during the entire life of the organism. Our research is aimed at understanding fundamental questions on cell proliferation control, cellular homeostasis and genome replication in multicellular organisms.
We have developed genomic strategies to study the functional properties and molecular determinants of DNA replication origins (ORIs) in all cell types of the whole organism to determine the influence of hormonal conditions, developmental signals and the environment (Fig. 1). We found that ORI activity is compatible with multiple signatures although most of them tend to associate with chromatin states present in proximal promoters, TSS and 5’-end of genes. In addition, ORIs are enriched in the tandem arrays of GGN trinucleotides, which can for G4 structures. Our experimental approach is opening a new avenue to use mutants in the analysis of genome replication. We are combining the study of molecular determinants of ORIs with detailed analysis of the pre-RC dynamics during organ development.
Cell proliferation is crucial for organogenesis, which is determined by a strict control of gene expression patterns. We study chromatin dynamics along the cell cycle with special emphasis in two aspects: one, the regulation of cell proliferation potential, very related to the control of gene expression in G1 and G2, and the exit to differentiation, and another, related to the specific chromatin modifications in response to stress. The balance between the canonical histone H3.1 and the variant H3.3 serves to identify the cell population undergoing their last cell cycle before exit to differentiation because most of H3.1 is massively evicted in the last G2 phase (Fig. 2), which is longer in these cells.
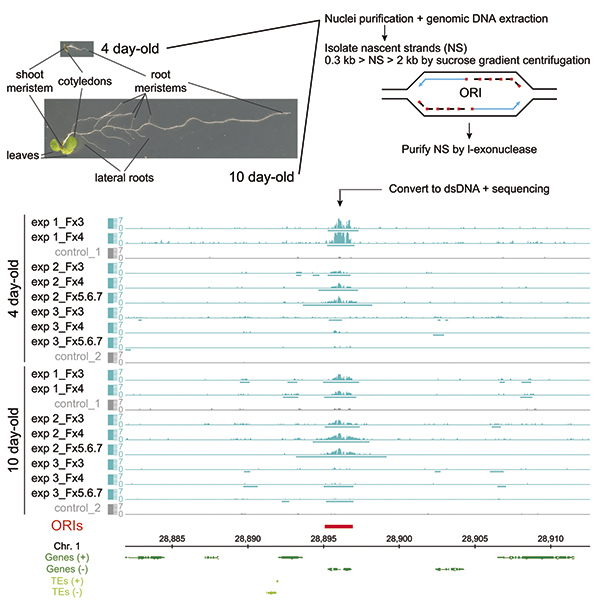
Fig. 1. Identification of DNA replication origins (ORIs) in whole developing organisms: 4 and 10 day-old Arabidopsis thaliana seedlings (left upper panel). The procedure includes the purification of single-stranded DNA of short nascent strands (SNS), conversion to double-stranded DNA and sequencing (right upper panel). Peak identification in several experimental situations allows the genome-wide mapping of ORIs (red in lower panel).
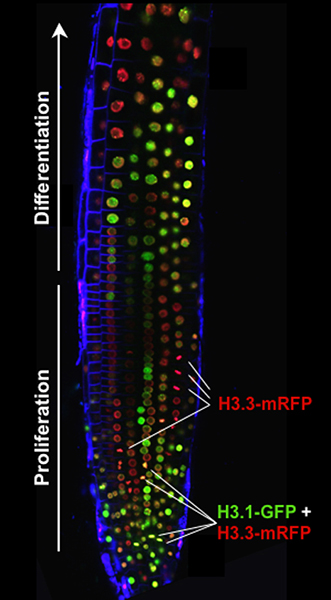
Fig. 2. Histone H3 dynamics. The canonical H3.1 (green), incorporated in every S-phase, but not the H3.3 variant (red), incorporated in a cell cycle-independent manner, is almost completely evicted in mitotic cells (red mitosis) undergoing their last division before entering the differentiation zone. Mitotic cells in previous cell divisions (yellow mitosis) contain both H3.1 and H3.3.

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Alonso Pérez | Elisa | 308 | 4658 | elisa.alonso(at)cbm.csic.es | E.Ayudantes De Invest. De Los Oo.Publicos De Investigacion |
| Desvoyes | Bénédicte | 308 | 4658 | bdesvoyes(at)cbm.csic.es | E.Científicos Titulares de Organismos Públicos de Investigación |
| Domínguez Pacheco | Juan Manuel | 308 | 4658 | Estudiante TFM | |
| Emiliani | Julia | 308 | 4658 | jemiliani(at)cbm.csic.es | M3 |
| Francisco Velilla | Mª del Rosario | 309 | 4649 | rfrancisco(at)cbm.csic.es | M3 |
| Gómez | María Sol | 308 | 4658 | sgomez(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Gómez Martínez | Diego | 308 | 4658 | diego.gomez(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Gutiérrez Armenta | Crisanto | 308 | 4638 | cgutierrez(at)cbm.csic.es | E. Profesores de Investigación de Organismos Públicos de Investigación |
| Hernández Sánchez Rebato | Miguel | 308 | 4638 | mhernandez(at)cbm.csic.es | M3 |
| López Gestoso | Miguel | 308 | 4658 | miguel.lopez(at)cbm.csic.es | M1 |
| Murugarren Garrido | Nerea | 308 | 4638 | nmurugarren(at)cbm.csic.es | M2 |
| Núñez Vázquez | Rocío | 308 | 4658 | rnunez(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Olmo Montoro | Iván | 308 | 4658 | iolmo(at)cbm.csic.es | Titulado Sup. Actividades Tecn. y Prof.GP1 |
| Pastor Solano | Adrián | 308 | 4658 | apastor(at)cbm.csic.es | M3 Predoc.formación |
Relevant publications:
- Han SK, Herrmann A, Yang J, Iwasaki R, Sakamoto T, Desvoyes B, Kimura S, Gutierrez C, Kim ED, Torii KU. Deceleration of the cell cycle underpins a switch from proliferative to terminal divisions in plant stomatal lineage. Dev Cell 57, 569-582 (2022)
- Sablowski, R., Gutierrez, C. Cycling in a crowd: coordination of plant cell division, growth and cell fate. Plant Cell 34, 193-208 (2022).
- Desvoyes B, Echevarría C, Gutierrez C. A perspective on cell proliferation kinetics in the root apical meristem.A perspective on cell proliferation kinetics in the root apical meristem. J Exp Bot. 72, 6708-6715 (2021)
- D’Ario, M., Tavares, R., Schiessl, K., Desvoyes, B., Gutierrez, C., Howard, M., Sablowski, R. Cell size controlled in plants using DNA content as an internal scale. Science 372, 1176–1181 (2021).
-
Echevarria, C., Gutierrez, C., Desvoyes, B. Tools for assessing cell cycle progression in plants. Plant Cell Physiol. 62, 1231-1238 (2021).
- Elena-Real, C., Gonzalez-Arzola, K., Diaz-Quintana, A., Velazquez-Campoy, A., Devoyes, B., Gutierrez, C., de la Rosa, M., Diaz-Moreno, I.
Cytochrome c modulates programmed cell death in plants as in humans by binding to 14-3-3 proteins. Plant J. 106, 74-85 (2021). - Gutierrez,C. Chromatin, DNA replication and transcription: closing the triangle. Trends Plant Sci. 26, 10-12 (2021).
- Desvoyes, B., Arana-Echarri, A., Barea, M.D., Gutierrez, C. A comprehensive fluorescent sensor for spatiotemporal cell cycle analysis in Arabidopsis. Nature Plants 6, 1330-1334 (2020).
- Probst, A., Desvoyes, B., Gutierrez, C. Similar yet Critically Different: The distribution, dynamics and function of Histone Variants. J. Exp. Bot. 71, 5191-5204 (2020) doi: 10.1093/jxb/eraa230.
- Desvoyes, B., Gutierrez, C. Roles of plant retinoblastoma protein: cell cycle and beyond. EMBO J. e105802 (2020).
- Ocaña-Pallarès, E., Vergara, Z., Desvoyes, B., Tejada-Jimenez, M., Romero-Jurado, A., Galvan, A., Fernandez, E., Ruiz-Trillo, I., Gutierrez, C. Origin recognition complex (ORC) evolution is influenced by global gene duplication / loss patterns in eukaryotic genomes. Genome Biol. Evol. (2020, in press, GBE-191130).
- Sequeira-Mendes, J., Vergara, Z., Peiró, R., Morata, J., Aragüez, I., Costas, C., Mendez-Giraldez, R., Casacuberta, J.M., Bastolla, U., Gutierrez, C. Differences in firing efficiency, chromatin and transcription underlie the developmental plasticity of Arabidopsis DNA replication origins. Genome Res. 29, 784–797 (2019).
- Desvoyes, B.*,+, Sequeira-Mendes, J.*, Vergara, Z., Madeira, S., Gutierrez, C+. Sequential ChIP protocol for profiling bivalent epigenetic modifications in the same chromatin fiber (ReChIP). * shared 1st coauthorship. Co-coresponding authors. Methods Mol. Biol. 1675, 83-97 (2017). In: Plant Chromatin Dynamics: methods and protocols. M. Bremer, C. Baroux, Eds. Springer Science.
- Vergara, Z., Sequeira-Mendes, J., Morata, J., Hénaff, E., Peiró, R., Costas, C., Casacuberta, J.M., Gutierrez, C. Retrotransposons are specified as DNA replication origins in the gene-poor regions of Arabidopsis heterochromatin. Nucleic Acids Res. 45, 8358–8368 (2017).
- Fernandez-Marcos, M., Desvoyes, B., Manzano, C., Liberman, L.M., Benfey, P.N., del Pozo, J.C., Gutierrez, C. Control of Arabidopsis lateral root boundaries by MYB36. New Phytol. 213, 105-112 (2017).
- Vergara, Z., Gutierrez, C. Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biol. 18, 96 (2017).
- Gutierrez, C., Desvoyes, B., Vergara, Z., Otero, S., Sequeira-Mendes, J. Links of genome replication, transcriptional silencing and chromatin dynamics. Curr. Opin. Plant Biol. 34, 92-99 (2016).
- Gutierrez, C. 25 years of cell cycle research: what’s ahead? Trends Plant Sci. 21, 823-833 (2016).
- Otero, S*., Desvoyes, B.*, Peiró, R., Gutierrez, C. Histone H3 Dynamics Reveal Domains with Distinct Proliferation Potential in the Arabidopsis Root. Plant Cell 28, 1361-1371 (2016) * shared 1st coauthorship. Plant Cell 28, 1235 (2016) In Brief- Hofmann, N. Last exit to differentiation: histone variants as signspots.
- Sequeira-Mendes, J., Aragüez, I., Peiró, R., Zhang, X., Jacobsen, S.E., Bastolla, U., Gutierrez, C. The Functional Topography of the Arabidopsis Genome is organized in a reduced Number of linear Motifs of Chromatin states. Plant Cell 26, 2351-2366 (2014).




















