Metabolism in cancer and aging
Diffuse large B cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults and represents a heterogeneous group of tumors with distinct subtypes that differ in genetic alterations, clinical outcome, response to treatment and prognosis. Approximately 5-15% of DLBCL are high grade B cell lymphomas (HGBL) with rearrangements of MYC and BCL2 and they are known as “double-hit” lymphoma (DHL) or “triple hit lymphoma” (THL) if they also present BCL6 translocations. Due to simultaneous activation of these driver oncogenes, DHLs are among the most aggressive and chemoresistant lymphoma subtype with minimal treatment options and have poor outcomes. Furthermore, the co-expression of MYC and BCL2 proteins without underlying rearrangements is considered a new adverse prognostic indicator termed double-expressor lymphoma (DEL). Deregulated MYC expression contributes to metabolic reprogramming in tumor cells through a complex network of factors. Specifically, c-MYC-transformed lymphoma B cells rely on glutamine metabolism for bioenergetics and redox homeostasis. In addition, serine metabolism, through the enhanced activity of SHMT2 enzyme, contributes to the biology of MYC-aggressive lymphomas. However, the metabolic features of the HGBL-DH lymphomas are not fully understood and could imply potentially targetable vulnerabilities. Understanding the unique biology of these tumors, including metabolism, could provide a rationale for exploring targeted agents beyond the current focus on BCL2 and MYC inhibitors.
Our lab, which opened its doors on September 2021, aims to characterize the metabolic features of these particular tumors using transcriptomic and metabololomic tools in preclinical models and patient samples, and screen new metabolic vulnerabilities that could be used as new targeted therapies for these aggressive lymphomas with very poor outcomes with the standard therapies.
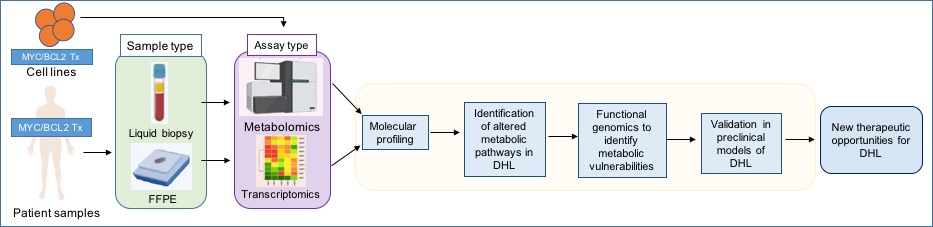
Figure 1. Research workflow

|
Apellidos |
Nombre |
Laboratorio |
Extensión |
|
Categoría Profesional |
|
Ortega Molina |
Ana |
408 |
4509 |
Investigador |
|
|
Jimeno Lumeras |
Rebeca |
408 |
4574 |
M3 | |
|
Prieto Carro |
Cristina |
408 |
4574 |
M2 | |
| López Martín-Lunas | Beatriz | 408 | 4574 | blopezml@cbm.csic.es | M2 |
| Salazar Ortego | Sara | 408 | 4574 | sara.salazaro@estudiante.uam.es | Beca JAE intro |
| González Pérez | Marta | 408 | 4574 | martgo78@ucm.es | Estudiante fin de máster |
1.- Ortega-Molina, A and Efeyan, A. (2021). From mouse genetics to targeting the Rag GTPase pathway. Molecular & Cellular Oncology 8 (5): 1979370. doi: 10.1080/23723556.2021.1979370
2.- Ortega-Molina, A*, Lebrero-Fernández, C., Sanz, A., Deleyto-Seldas, N, Plata-Gómez A.B., Menéndez, C., Graña-Castro O., Caleiras, E., Efeyan, A*. (2021). Inhibition of Rag GTPase signaling in mice suppresses B cell responses and lymphomagenesis with minimal detrimental tradeoffs. Cell Reports, 36(2):109372. doi: 10.1016/j.celrep.2021.109372
3.- De la Calle Arregui C., Plata-Goméz A.B., Deleyto-Seldas N., García F., Ortega-Molina A., Abril-Garrido J., Rodriguez E., Nemazanyy I., Tribouillard L., de Martino A., Caleiras E., Campos-Olivas C., Mulero M., Laplante M., Muñoz J., Pende M., Sabio G., Sabatini D.M., Efeyan A. (2021). Limited survival and impaired hepatic fasting metabolism in mice with constitutive Rag GTPase signaling. Nature Communications, 12(1):3660. doi: 10.1038/s41467-021-23857-8
4.- Parsa, S#., Ortega-Molina, A#, Ying HY., Jiang, M., Teater, M., Wang, J., Zhao, C., Reznik, E., Pasion, JP., Kuo, D., Mohan, P., Wang, S., Camarillo, JM., Thomas, PM., Jain, N., Garcia-Bermudez, J., Cho, B., Tam, W., Kelleher, NL., Socci, N., Dogan, A., De Stanchina, E., Ciriello, G., Green, M., Li, S., Birsory, K., Melnick, AM., Wendel, HG. (2020). The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nature Cancer, 1: 653-664. doi: 10.1038/s43018-020-0080-0
5.- Ortega-Molina A., Deleyto-Seldas N., Carreras J. Sanz, A., Lebrero-Fernandez, C., Menendez, C., Vandenberg, A., Fernandez-Ruiz, B., Marin-Arraiza, L., de la Calle Arregui, C., Plata-Gomez, AB., Caleiras, E., de Martino, A., Martinez-Martin, N., Troule, K., Piñeiro-Yañez, E., Nakamura, Y., Araf, S., Victora, GD., Okosun, J., Fitzgibbon, J., Efeyan, A. (2019). Oncogenic Rag GTPase ignaling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Nature Metabolism, 1 (8): 775-789. doi: 10.1038/s42255-019-0098-8
6.- Jiang Y#, Ortega-Molina A#, Geng H#, Ying HY#, Hatzi K, Parsa S, McNally D, Wang L, Doane AS, Agirre Ena X, Teater M, Meydan C, Li Z, Poloway D, Wang S, Ennishi D, Scott DW, Stengel KR, Kranz JE, Holson E, Sharma S, Young JW, Chu CS, Roeder RG, Shaknovich R, Hiebert SW, Gascoyne RD, Tam W, Elemento O, Wendel HG, Melnick AM. (2017). CREBBP Inactivation Promotes the Development of HDAC3 Dependent Lymphomas. Cancer Discovery, 7(1): 38-53. doi: 10.1158/2159-8290.CD-16-0975
7.- Ortega-Molina A.#, Boss I. W.#, Canela A.#., Pan H., Jiang Y., Zhao C., Jiang M., Hu D., Agirre X., Niesvixky I., Lee J.-E., Chen H.-T., Ennishi D., Scott D., Mottok A., Hother C., Liu S., Cao X.-J., Tam W., Shaknovich R., Garcia B. A., Gascoyne R., Ge K., Shilatifard A., Elemento O., Nussenzweig A., Melnick A. M., Wendel H.G. (2015). The histone lysisne methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nature Medicine, 21(10): 1199-1208. doi: 10.1038/nm.3943
*Co-corresponding authors
# Co-first authors




















