Genetic variability of RNA viruses
Research summary:
Our group pioneered (1978-1990) the discovery of viral quasispecies, a highly complex and dynamic population structure, with profound implications in viral evolution and pathogenesis. Our results of the last 40 years on quasispecies dynamics have been amply confirmed by application of deep sequencing methodology in several laboratories in the last years. The genome of an RNA viral population is a weighted average of myriads of different, related sequences which are changing continuously. The consensus (or average) sequence that we write for convenience, and that fills textbooks and data banks, is an abstraction that may not even exist in the population that the sequence aims at representing. We have recently obtained evidence of a new class of positive selection that we term “broadly diversifying selection” that provides a very large repertoire of viral genomes with similar replicative fitness despite a history of prolonged viral replication in the same cellular environment. Dynamic mutant clouds are prepared to respond to changing environments by selection of subsets of sequences preferentially over others, an event crucial to disease progression. We are currently attempting to link evolutionary events at the quasispecies (intra-host) and epidemiological (inter-host) levels, important for the design of effective antiviral designs.
Quasispecies dynamics demands that new approaches be investigated for the prevention and treatment of diseases associated with RNA viruses, to counteract the adaptive capacity conferred by the mutant clouds.
We are working in the development of new antiviral strategies that can avoid selection of treatment-escape mutant viruses. The work involves several pathogenic viruses, with the hepacivirus hepatitis C virus (HCV) as our main model. We are collaborating with several groups to expand the scope of our work: with the group of Nuria Verdaguer (IBM, CSIC); Pablo Gastaminza (CNB, CSIC); Celia Perales (IIS, Fundación Jiménez Díaz); Josep Quer (Hospital Vall d’Hebrón); Aurora Sánchez (IIB, CSIC-UAM); Juan Carlos Saiz and Alejandro Brun (INIA).
During the last years, our emphasis has been on HCV given its magnitude as public health problem worldwide, and the direct implication of its quasispecies nature in the hepatic disease and response to treatments. The main contributions of our group to HCV have been: (i) that interferon resistance is multigenic; (ii) to make available HCV populations of different fitness level to study fitness effects on HCV biology; (iii) that viral fitness is a determinant of multi-drug resistance in absence of specific resistance mutations, (iv) the absence of population equilibrium (steady-state distribution of mutations and phenotypic stasis) even after extensive multiplication in a constant cellular environment, and (v) the design of sequential and combination treatments using both mutagenic and non-mutagenic antiviral agents for suppression of viral infectivity (summary in Figure 1). These investigations continue at the present time. The collaboration with the group of Dr. Celia Perales is essential to connect results with HCV in cell culture and observations with infected patients.
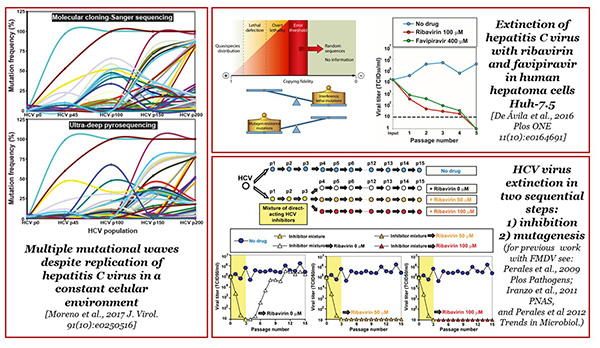
Figure 1. Graphic summary of our recent work on quasispecies implications, and lethal mutagenesis of RNA viruses. The left panel illustrates HCV mutational waves (color-coded) quantified by molecular cloning-Sanger sequencing and deep sequencing during viral multiplication in a constant biological environment. The waves drawn here have been found by the two methods, excluding that they are due to a sampling bias. Panels on the right illustrate the conceptual basis of lethal mutagenesis, with an example of HCV extinction (top), a serial passage design (middle), and HCV extinction associated with ribavirin treatment (bottom). Detailed information is in the indicated references, and the reference list.
Lethal mutagenesis is an expanding field of research that has opened a new chapter in antiviral pharmacology. Our work, in collaboration with the late John Holland (UCSD), provided the first experimental evidence that an increase of mutation rate promoted by chemical mutagenesis was detrimental to RNA virus survival (1990-1997). In our current screening of mutagenic agents that have been licensed for human use, we have documented that the broad spectrum antiviral agent favipiravir (T-705) is mutagenic for FMDV and HCV, and can lead to the extinction of these viruses. We have collaborated with the groups of Juan Carlos Saiz and Alejandro Brun (INIA) to show that favipiravir is also mutagenic for West Nile and Rift Valley fever viruses, and can drive the virus towards extinction in cell culture. We have recently documented the molecular basis of a synergistic antiviral activity of favipiravir and ribavirin on HCV, which is promising for the control of emerging and reemerging viral infections. A major unresolved issue is the integration of mutagenic and non-mutagenic inhibitors into antiviral designs to combat RNA viral diseases.
We have discovered a new mechanism of resistance to lethal mutagenesis of FMDV to ribavirin and 5-fluorouracil, consisting in selection of viral mutants that can modulate nucleotide incorporation to avoid the mutational bias produced by mutagenic nucleotides. This mechanism does not alter the amplitude of the mutant spectrum, and the corresponding adaptive flexibility. Interestingly, a joker mutation (meaning a given mutation that is repeatedly encountered when the virus is subjected to different selective constraints) in non-structural protein 2C modulated nucleotide incorporation in response to ribavirin mutagenesis, in absence of mutations in the viral polymerase. These observations emphasize the multiple adaptive resources that RNA viruses have to respond to hostile environments, and justify our ongoing efforts to search for antiviral designs to strongly suppress virus multiplication.
In an interesting connection between virus expansion in the field and the clonal evolution of organisms, we have proposed for viruses a distinction between mechanistically active but inconsequential recombination, and evolutionary relevant recombination. Based on the available evidence, we think that mutation and recombination occur continuously during RNA virus replication, but that a majority of mutant and recombinant genomes do not survive (they are subjected to negative selection). However, there are subsets of mutations that combined with punctuated, biologically relevant recombination events, contribute to virus survival and evolution (Figure 2). These studies are contributing also to a deeper understanding of the molecular events underlying viral disease processes.
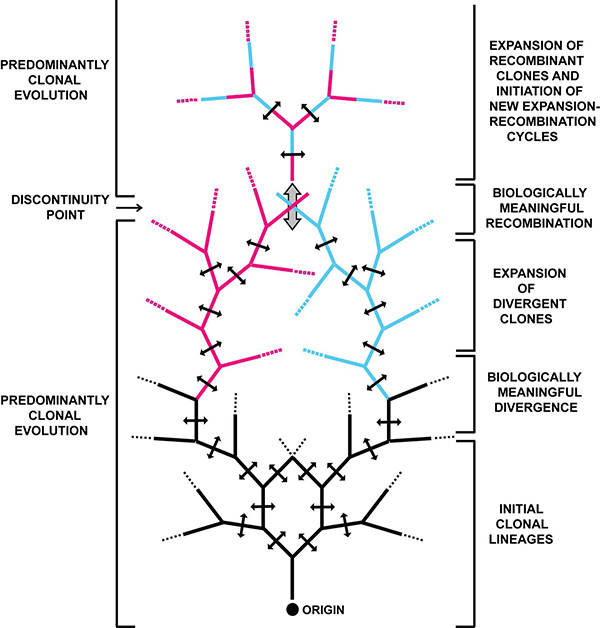
Figure 2. Schematic representation of clonal evolution of viruses. From an initial infection (Origin, bottom) multiple sublineages are generated. Recombination takes place at any branch (double-headed arrows perpendicular to branches). Biologically relevant diversification is illustrated by red and blue branches. Recombination at a discontinuity point (large vertical double-headed arrow) is biologically relevant because it generates mosaic genomes with new phenotypes. Clonal evolution continues until a new discontinuity point is reached. The scheme does not imply a space or time scale. Reproduced from Perales et al. 2015, with permission from the National Academy of Sciences USA.
Our major aim is to use the new understanding of viral dynamics that has been provided by the framework of quasispecies theory to find new ways to suppress viral multiplication. It is our conviction that new antiviral designs, which are based on the understanding of the implications of quasispecies, can be used to control established, emergent, and re-emergent viral pathogens (Figure 3).
Figure 3. A summary of the approach and main goal of our research.

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Domingo Soláns | Esteban | 121 | 4540 | edomingo(at)cbm.csic.es | Doctor Vinculado "Ad Honorem" |
| Somovilla Crespo | Pilar | 121 | 4541 | psomovilla(at)cbm.csic.es | Investigador Doctor |
Relevant publications:
- Ojosnegros, S., Beerenwinkel, N., Antal, T., Nowak, M.A., Escarmís, C. and Domingo, E. (2010). Competition-colonization dynamics in an RNA virus. Proc. Natl. Acad. Sci. USA., 107, 2108-2112.
- Agudo, R., Ferrer-Orta, C., Arias, A., de la Higuera, I., Perales, C., Pérez-Luque, R., Verdaguer, N. and Domingo, E. (2010) A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape. PLoS Pathog., 6(8): e1001072.
- Ojosnegros, S., García-Arriaza, J., Escarmís, C. Manrubia, S.C., Perales, C., Arias, A., García-Mateu, M. and Domingo, E. (2011). Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genetics, 7(3): e1001344.
- Moreno, E., Ojosnegros, S., García-Arriaza, J., Escarmís, C., Domingo, E. and Perales C. (2014). Exploration of sequence space as the basis of viral RNA genome segmentation. Proc Natl Acad Sci USA, 111(18): 6678-6683.
- De Ávila, A.I., Gallego, I., Soria, M.E., Gregori, J., Quer, J., Esteban, J.I., Rice, C.M., Domingo, E., Perales, C. (2016). Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS ONE, 11 (10): e0164691.
- Moreno, E.; Gallego, I.; Gregori, J.; Lucia-Sanz, A.; Soria, M. E.; Castro, V.; Beach, N. M.; Manrubia, S.; Quer, J.; Esteban, J. I.; Rice, C. M.; Gomez, J.; Gastaminza, P.; Domingo, E.; Perales, C. (2017). Internal Disequilibria and Phenotypic Diversification during Replication of Hepatitis C Virus in a Noncoevolving Cellular Environment. J. Virol., 91 (10), e02505-16.
- de la Higuera, I.; Ferrer-Orta, C.; de Avila, A. I.; Perales, C.; Sierra, M.; Singh, K.; Sarafianos, S. G.; Dehouck, Y.; Bastolla, U.; Verdaguer, N.; Domingo, E. (2017). Molecular and Functional Bases of Selection against a Mutation Bias in an RNA Virus. Genome Biol. Evol., 9 (5), 1212-1228.
- De la Higuera, I., Ferrer-Orta, C., Moreno, E., De Avila, A.I., Soria, M.E., Singh, K., Caridi, F., Sobrino, F., Sarafianos, S.G., Perales, C., Verdaguer, N., and Domingo, E. (2018). Contribution of a multifunctional polymerase region of foot-and-mouth disease virus to lethal mutagenesis. J. Virol. 92(20). pii: e01119-18.
- Gallego, I., Soria, M.E., Gregori, J., de Ávila, García-Crespo, C., Moreno, E., Gadea, I., Esteban, J., Fernández-Roblas, R., Esteban, J.I., Gómez, J., Quer, J., Domingo, E., Perales, C. (2019). Synergistic lethal mutagenesis of hepatitis C virus. Antimicrob Agents Chemother. pii: AAC.01653-19. doi: 10.1128/AAC.01653-19.
- Gallego, I., Soria, M.E., García-Crespo, C., Chen, Q., Martínez-Barragán, P., Khalfaoui, S., Martínez-González, B., Sanchez-Martin, I., Palacios-Blanco, I., de Ávila, A.I., García-Cehic, D., Esteban, J.I., Gómez, J., Briones, C., Gregori, J., Quer, J., Perales, C., Domingo, E. (2019). Broad and dynamic diversification of infectious hepatitis C virus in a cell culture environment. J.Virol. JVI.01856-19; doi: 10.1128/JVI.01856-19.
Doctoral theses:
He has directed or co-directed 26 Doctoral Theses. They are listed only beginning in 2012.
- Héctor Moreno Borrego (2012). Dinámica poblacional del virus de la coriomeningitis linfocitaria del ratón en su interacción con agentes mutagénicos. Universidad Autónoma de Madrid. Directores: Esteban Domingo y Verónica Martín.
- Ignacio de la Higuera Hernández (2014).Factores determinantes del reconocimiento de nucleótidos en el virus de la fiebre aftosa. Universidad Autónoma de Madrid. Director: Esteban Domingo.
- Ana Mª Ortega Prieto (2014).Mutagénesis letal del virus de la Hepatitis C. Universidad Autónoma de Madrid. Directores: Esteban Domingo y Celia Perales.
- Elena Moreno del Olmo(2017). Evolución a largo plazo de virus RNA en ambiente biológico constante. Universidad Autónoma de Madrid. Directores: Esteban Domingo y Celia Perales.
Patents:
- N. Sevilla, E. Domingo, C. Escarmís, S. Ojosnegros, J. García-Arriaza, M. Sanz-Rojo, T. Rodríguez. "Vacuna atenuada para la fiebre aftosa". Nº DE SOLICITUD: P200801583. Patente concedida en España el 16/06/2011. Nº PUB: ES2344875.
Other activities:
- International member of National Academy of Sciences (USA), (2020).
- Académico Numerario de la Real Academia de Ciencias Exactas, Físicas y Naturales, adscrito a la Sección de Ciencias Naturales, (2011).
- Miembro de la Red Española de Biofísica, coordinada por el Dr. David Reguera, desde 2011.
- Miembro del Global Virology Network, coordinado por el Dr. Robert Gallo, desde 2011.
- Miembro del Comité Organizador del Congreso FEMS 2011 (Ginebra, Suiza, 2011).
Ongoing projects:
- Estudio de las secuelas moleculares en la célula hospedadora tras la erradicación del virus de la hepatitis C en cultivo celular. SAF2017-87846-R. Financiado por: Ministerio de Ciencia, Innovación y Universidades. Investigador Principal: Pablo Gastaminza y Esteban Domingo. (2018-2020). Convocatoria: PN2017 - Programa estatal de i+d+i orientada a los retos de la sociedad - Plan estatal de investigación científica y técnica y de innovación 2013-2016.
- Plataforma para el desarrollo de estrategias de control de salud animal. PLATESA2, S2018/BAA-4370. Financiado por: Comunidad de Madrid/FEDER. Coordinador: Noemí Sevilla. IP grupo VIRNA: E. Domingo. (2019-2022). Convocatoria: CM -Convocatoria de ayudas para la realización de programas de actividades de I+D entre grupos de investigación de la Comunidad de Madrid (CM) en Tecnologías 2018.
- Búsqueda de cDNA del virus de la hepatitis C. BFU2017-91384-EXP. Financiado por: Ministerio de Ciencia, Innovación y Universidades. Investigador Principal: Celia Perales. (2018-2020). Convocatoria: PN2017 – Proyecto Explora - Subprograma estatal de generación de conocimiento - Programa estatal de fomento de la investigación científica y técnica de excelencia 2013-2016.
- Combinaciones de antivirales frente al SARS-CoV-2. CSIC-COV19-014. Financiado por: Agencia Estatal CSIC. Investigador Principal: Nuria Verdaguer. (2020-2021). Convocatoria: Ayudas CSIC-COVID-19.




















