Molecular modelling group
Research summary:
Computational biology lab. The work is devoted to the integration of evolutive and structural information to study the function of proteins, the simulation of dynamic processes of protein-protein and protein-ligand interaction, the development of novel "in silico" drug design systems and the generation of new quantitative methods for computational biology.
Current projects:
- A. Analysis by computational simulation of enzymatic reactions catalyzed by enzymes of biomedical interest. Design of specific inhibitors.- Cohesin: Analysis of the molecular interactions among the protein components of the cohesin ring and the interaction of the protein complex with DNA
-
- - FtsZ: Simulation by molecular dynamics of the processes of polymerization and depolymerization of the bacterial septum protein FtsZ, associated with the GTPase activity of its active center.
- - Carbapenemases. Dynamic simulation of the interaction of bacterial carbapenemases (VIM-2, KPC-2, OXA-48) with substrates and known inhibitors.
- B. Development of a new and efficient drug design system based on computational dynamic simulation of macromolecular structures. Based on the analysis of enzyme active centers, the method developed by our group consists of simulating these protein structures through molecular dynamics for several hundred nanoseconds, selecting different representative structures and filtering a database of 3D compounds for each one of them:
-
- - Molecules able to inhibits the cell cycle of human tumor cells, with the potential to be used as an anti-tumor drugs.
- - Bacterial cell division cycle inhibitor molecules that can be used as potential antimicrobials.
- - New carbapenemases inhibitors ready to enter into clinical trials.
- C. Development of a new and efficient method (MEPSAnd) that natively calculates minimum energy paths across energy surfaces of any number of dimensions.
Group website: http://www.cbm.csic.es/bioweb
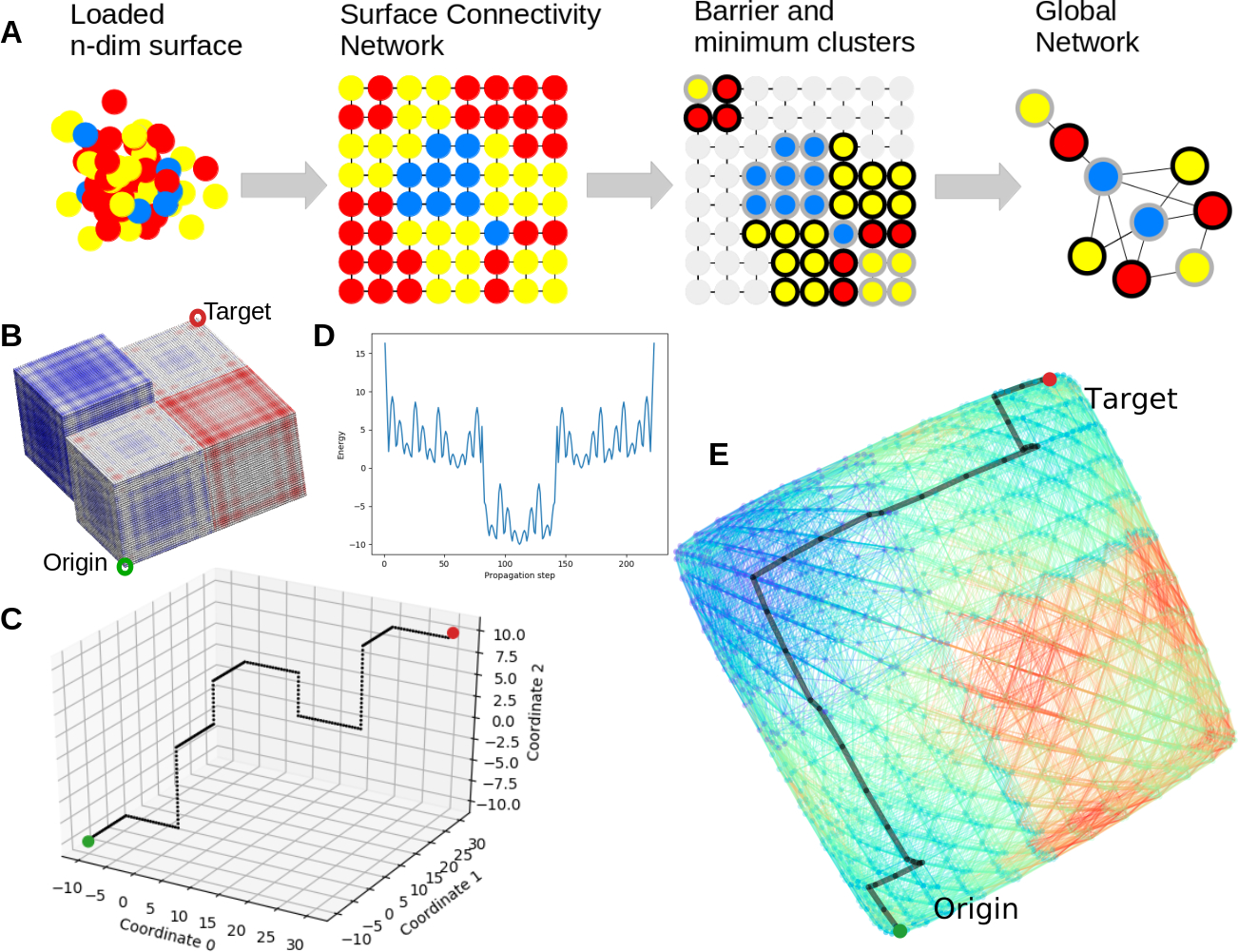
Figure 1. Overview of MEPSAnd, a new and efficient method developed in the group that natively calculates minimum energy paths across energy surfaces of any number of dimensions. The figure shows a diagram of the MEPSAnd algorithm steps and some examples and applications [doi: 10.1093/bioinformatics/btz649].

| Last name | Name | Laboratory | Ext.* | Professional category | |
|---|---|---|---|---|---|
| Gómez Puertas | Paulino | 312 | 4663 | pagomez(at)cbm.csic.es | E.Científicos Titulares de Organismos Públicos de Investigación |
| Marcos Alcalde | Iñigo | 312 | 4662 | imarcos(at)cbm.csic.es | M3 66,66% |
| Ros Pardo | David | 313.1 | 4662 | davidrp(at)cbm.csic.es | M3 |
Recent publications:
- Marcos-Alcalde, I., Lopez-Viñas, E. & Gómez-Puertas, P. (2020). MEPSAnd: Minimum Energy Path Surface Analysis over n-dimensional surfaces. Bioinformatics 36, 956–958. doi:1093/bioinformatics/btz649
- Latorre-Pellicer, A., Ascaso, A., Trujillano, L., Gil-Salvador, M., Arnedo, M., Lucia-Campos, C., Antoñanzas-Pérez, R., Marcos-Alcalde, I., Parenti, I., Bueno-Lozano, G., Musio, A., Puisac, B., Kaiser, F.J., Ramos, F.J., *Gómez-Puertas, P. & *Pié, J. (*Corresponding authors) (2020). Evaluating Face2Gene as a Tool to Identify Cornelia de Lange Syndrome by Facial Phenotypes. International Journal of Molecular Sciences 21, 1042. doi:3390/ijms21031042
- Lazo, P.A., García, J.L., Gómez-Puertas, P., Marcos-Alcalde, I., Arjona, C., Villarroel, A., González-Sarmiento, R. & Fons, C. (2020). Novel dominant KCNQ2 exon 7 partial in-frame duplication in a complex epileptic and neurodevelopmental delay syndrome. International Journal of Molecular Sciences 21, 4447. doi:3390/ijms21124447
- Marcos, A.T., Martín-Doncel, E., Morejón-García, P., Marcos-Alcalde, I., Gómez-Puertas, P., Segura-Puimedon, M., Armengol, L., Navarro-Pando, J.M. & Lazo, P.A. (2020). VRK1 (Y213H) homozygous mutant impairs Cajal bodies in a hereditary case of distal motor neuropathy. Annals of Clinical and Translational Neurology 7, 808-818. doi:1002/acn3.51050
- Ruiz-Márvez, E., Ramírez, C.A., Rodríguez, E.R., Flórez, M.M., Delgado, G., Guzmán, F., Gómez-Puertas, P., Requena, J.M. & Puerta, C.J. (2020). Molecular characterization of Tc964, a novel antigenic protein from Trypanosoma cruzi. International Journal of Molecular Sciences 21, 2432. doi:3390/ijms21072432
- Krab, L.C., Marcos-Alcalde, I., Assaf, M., Balasubramanian, M., Andersen, J.B., Pedersen, A-M.B., Cefle, K., Fitzpatrick, D., Gudmundsson, S., Huisman, S., McKee, S., Maas, S.M., Menke, L.A., Mulder, P.A., Martínez, F., Mokry, J., Murch, O.D., Parker, M., Pie, J., Ramos, F., Rieubland, C., Scarano, E., Shinawi, M., Gómez-Puertas, P., Tümer, Z. & Hennekam, R.C. (2020). Delineation of phenotypes related to cohesin structural protein RAD21. Human Genetics 139, 575–592. doi:1007/s00439-020-02138-2
- Arnedo, M., Latorre-Pellicer, A., Lucia-Campos, C., Gil-Salvador, M., Antoñanzas-Pérez, R., Gómez-Puertas, P., Bueno-Lozano, G., Puisac, B. & Pié, J. (2019). More Than One HMG-CoA Lyase: The Classical Mitochondrial Enzyme Plus the Peroxisomal and the Cytosolic Ones. International Journal of Molecular Sciences 20, 6124. doi:3390/ijms20246124
- Gudmundsson, S., Annéren, G., Marcos-Alcalde, I., Wilbe, M., Melin, M., Gómez-Puertas, P. & Bondeson, M-L. (2019). A novel RAD21 p.(Gln592del) variant expands the clinical description of Cornelia de Lange syndrome type 4 - review of the literature. European Journal of Medical Genetics 62, 103526. doi:1016/j.ejmg.2018.08.007
- Reis, F.P., Bárria, C., Gómez-Puertas, P., Gomes, C.M. & Arraiano, C.M. (2019). Identification of temperature-sensitive mutations and characterization of thermolabile RNase II variants. FEBS Letters 593, 352-360. doi:1002/1873-3468.13313
- Marcos-Alcalde, I., Mendieta-Moreno, J.I., Puisac, B., Gil-Rodríguez, M.C., Hernández-Marcos, M., Soler-Polo, D., Ramos, F.J., Ortega, J., Pié, J., Mendieta, J. & Gómez-Puertas, P. (2017). Two-step ATP-driven opening of cohesin head. Scientific Reports 7, 3266. http://dx.doi.org/10.1038/s41598-017-03118-9
- Marcos-Alcalde, I., Setoain, J., Mendieta-Moreno, J.I., Mendieta, J. & Gómez-Puertas, P. (2015). MEPSA: minimum energy pathway analysis for energy landscapes. Bioinformatics 31, 3853-3855. http://dx.doi.org/10.1093/bioinformatics/btv453
- Mendieta-Moreno, J., Walker, R., Lewis, J., *Gómez-Puertas, P., Mendieta, J. & *Ortega, J. (*Corresponding authors) (2014). FIREBALL/AMBER: An efficient local-orbital DFT QM/MM method for biomolecular systems. Journal of Chemical Theory and Computation 10, 2185–2193. http://dx.doi.org/10.1021/ct500033w
- Martín-García, F., Mendieta-Moreno, J.I., Marcos-Alcalde, I, *Gómez-Puertas, P. & Mendieta, J. (*Corresponding author). (2013). Simulation of catalytic water activation in mitochondrial F1-ATPase using a hybrid quantum mechanics/molecular mechanics approach: An alternative role for ß-Glu 188. Biochemistry 52, 959-966. http://dx.doi.org/10.1021/bi301109x




















